con <- dbConnect(odbc::odbc(),
Driver = ".../lib/libsparkodbc_sb64-universal.dylib",
Host = Sys.getenv("DATABRICKS_HOST"),
PWD = Sys.getenv("DATABRICKS_TOKEN"),
HTTPPath = "/sql/1.0/warehouses/300bd24ba12adf8e",
Port = 443,
AuthMech = 3,
Protocol = "https",
ThriftTransport = 2,
SSL = 1,
UID = "token")Welcome to
Using Databricks
with R
Workshop
August 12
Housekeeping items
- posit::conf mobile app
- Wi-fi password
- Accessing content -pos.it/databricks_24
Schedule
9am - 10:30am
Break (30mins)
11am - 12:30pm
Lunch (1hr)
1:30pm - 3pm
Break (30mins)
3:30pm - 5pm
The team


Edgar Ruiz
Instructor
James Blair
TA
Materials
- Posit Workbench Server
- Databricks Cluster
- Deck
- Exercise book
Posit + Databricks
Special work we have done as part of the new partnership

Let’s get started…
Log into Posit Workbench
Step 1 - Landing Page
Navigate to conf.posit.team, click on Posit Workbench

Step 2 - OpenID Page
Click on Sign in with OpenID
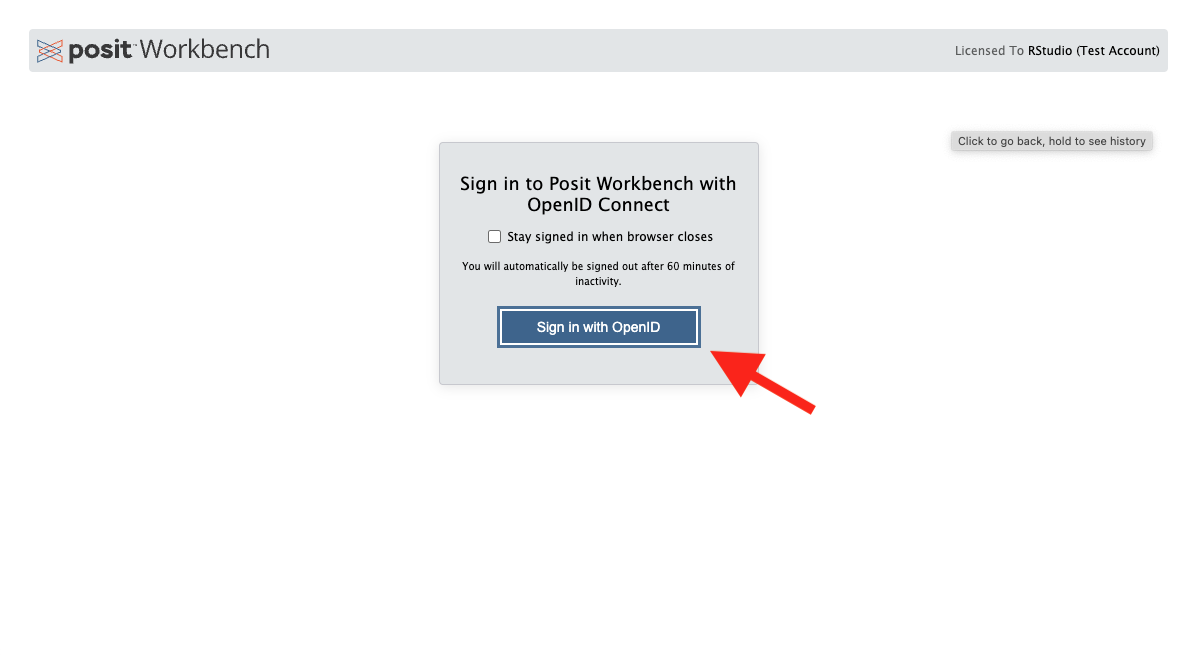
Step 3 - KeyCloak Page
Click on the GitHub icon
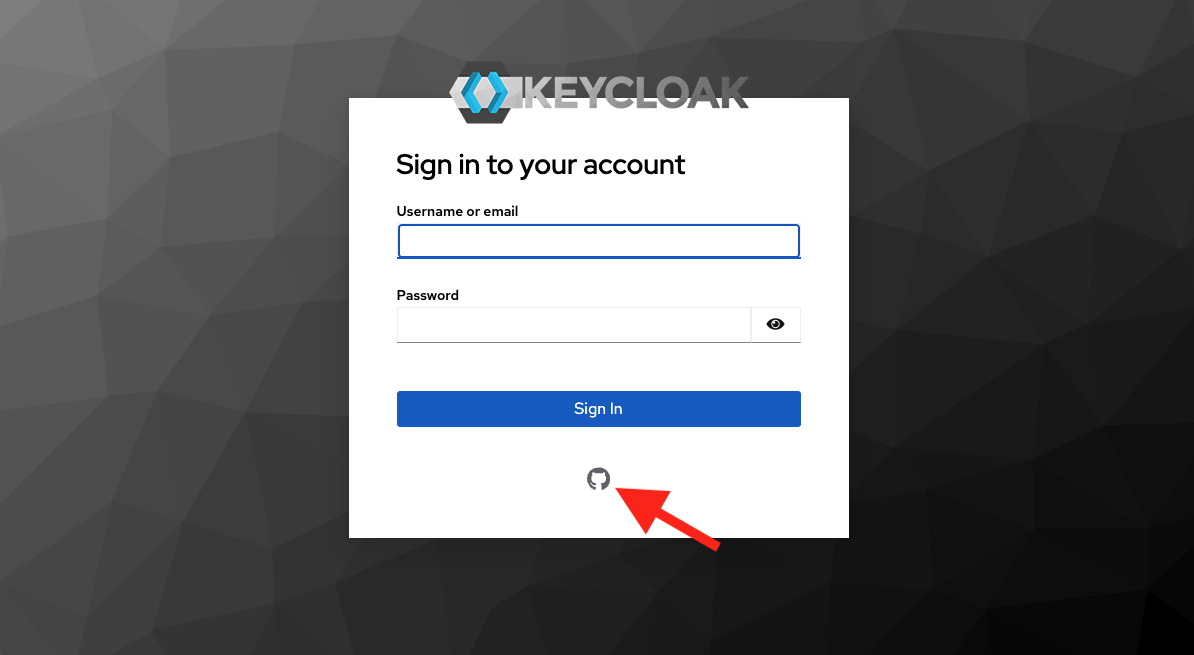
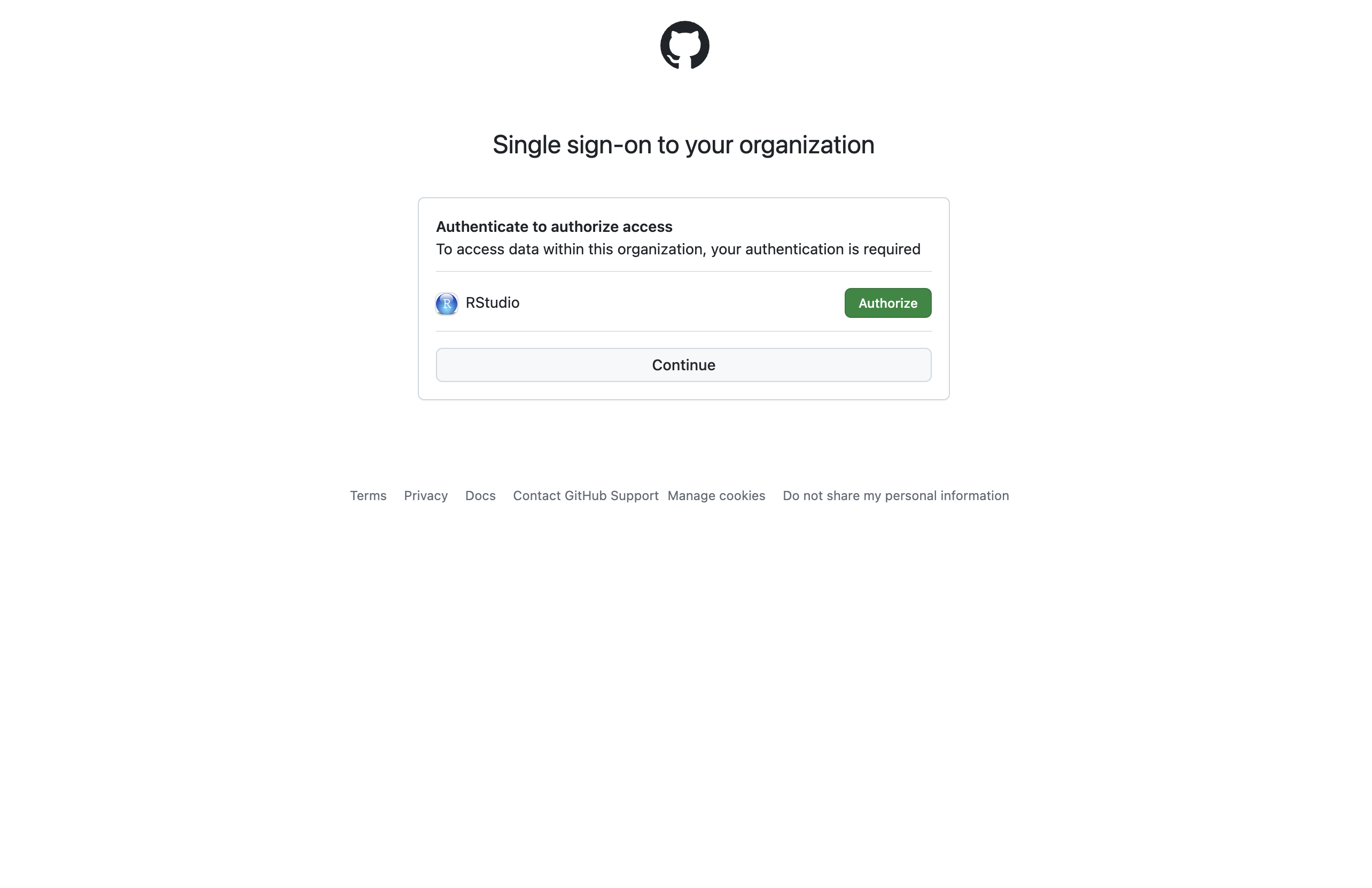
Step 4 - GitHub
Log into GitHub, and/or approve the RStudio org
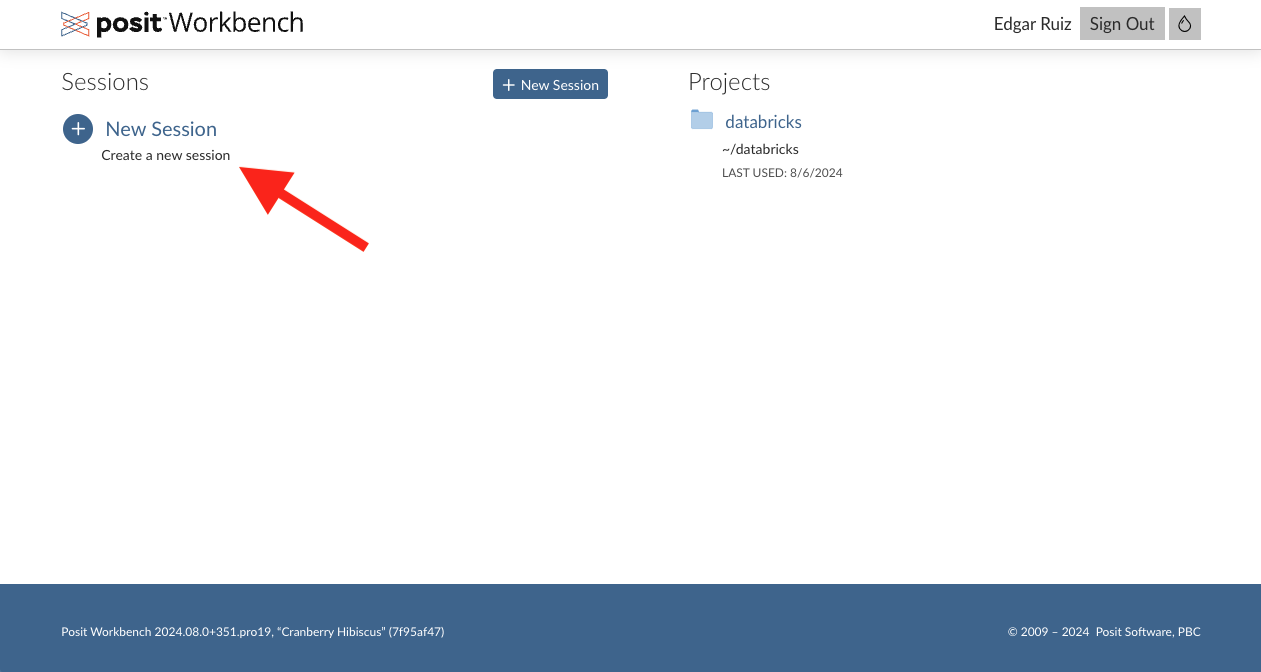
Step 5 - Posit Workbench homepage
Click on the New Session button
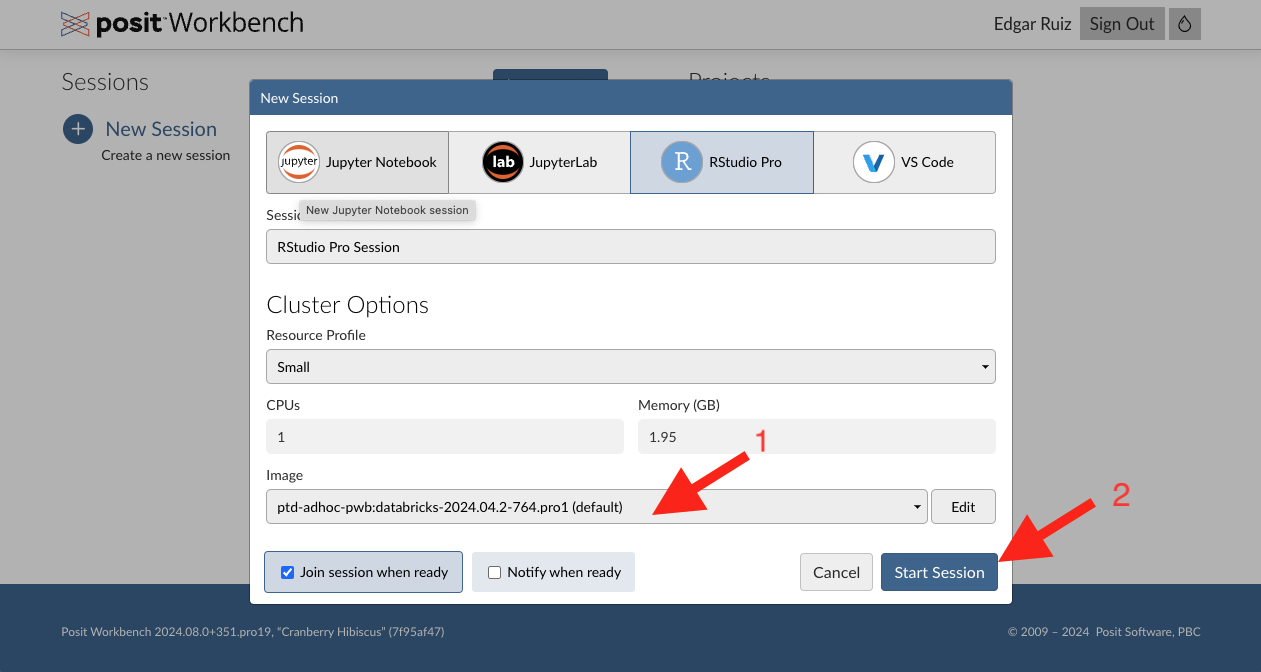
Step 6 - Setup new session
Confirm that the image matches what’s on the screenshot (1), then click on Start Session (2).
Step 7 - Run setup command
In the R console type
“Embrace the remoteness”
Martin Grund, Databricks
Key concept
🔑 Data & processing need to be as physically close as possible
Working with Databricks
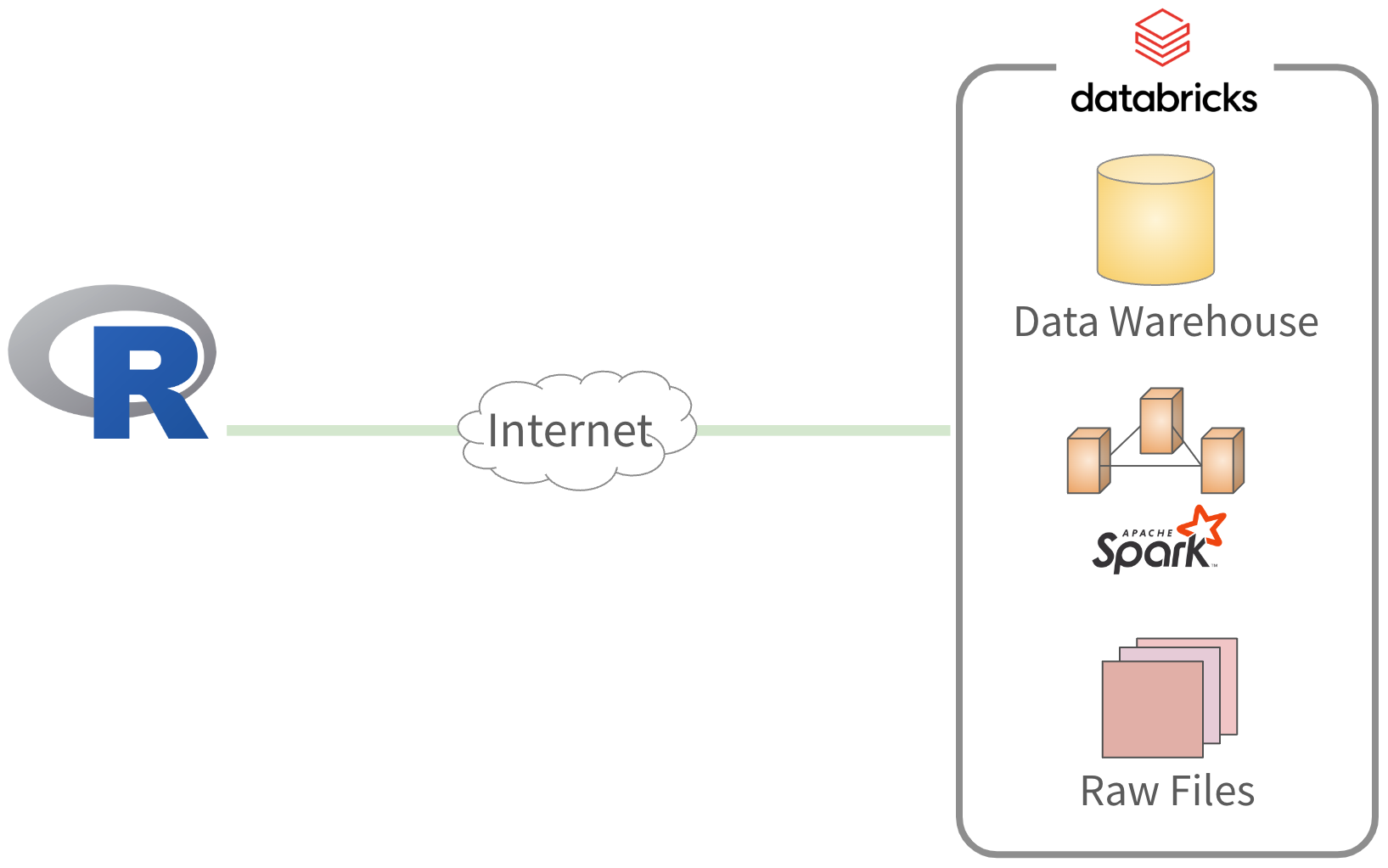
“Default” approach



“Default” approach




“Default” approach





Better approach!



Better approach!





Better approach!






Takeaways
- Download data if doing something only R can do
- 🔑 Data & processing need to be as physically close as possible
- Move most of the processing to Databricks
- But how?…
Unit 1
Accessing
Databricks
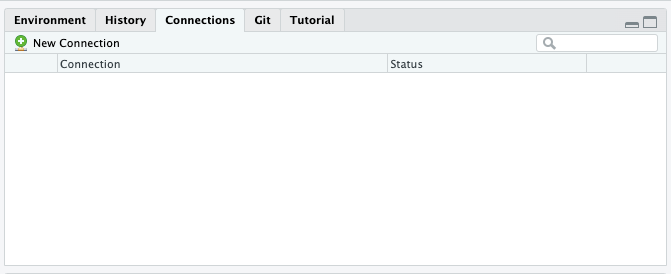
Using RStudio
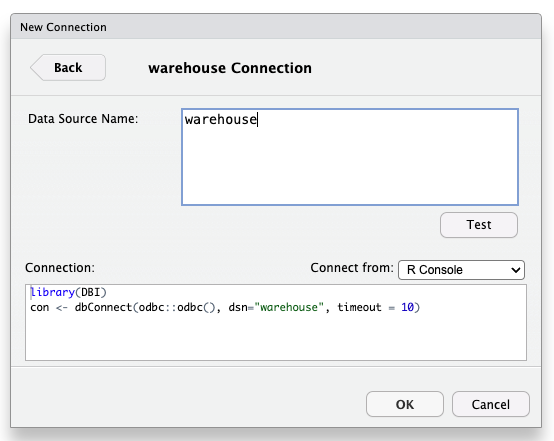
In the “Connections” pane, select “New Connection”
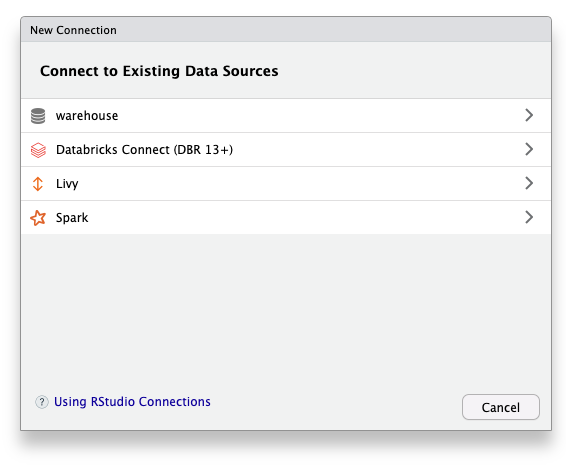
Using RStudio
Select ‘warehouse’
Using RStudio
Click ‘OK’
RStudio DB Navigator
Explore the catalogs, schema and tables

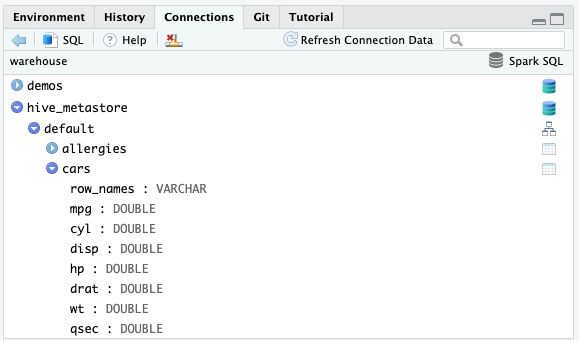
Exercises 1.1 - 1.2
How everything connects
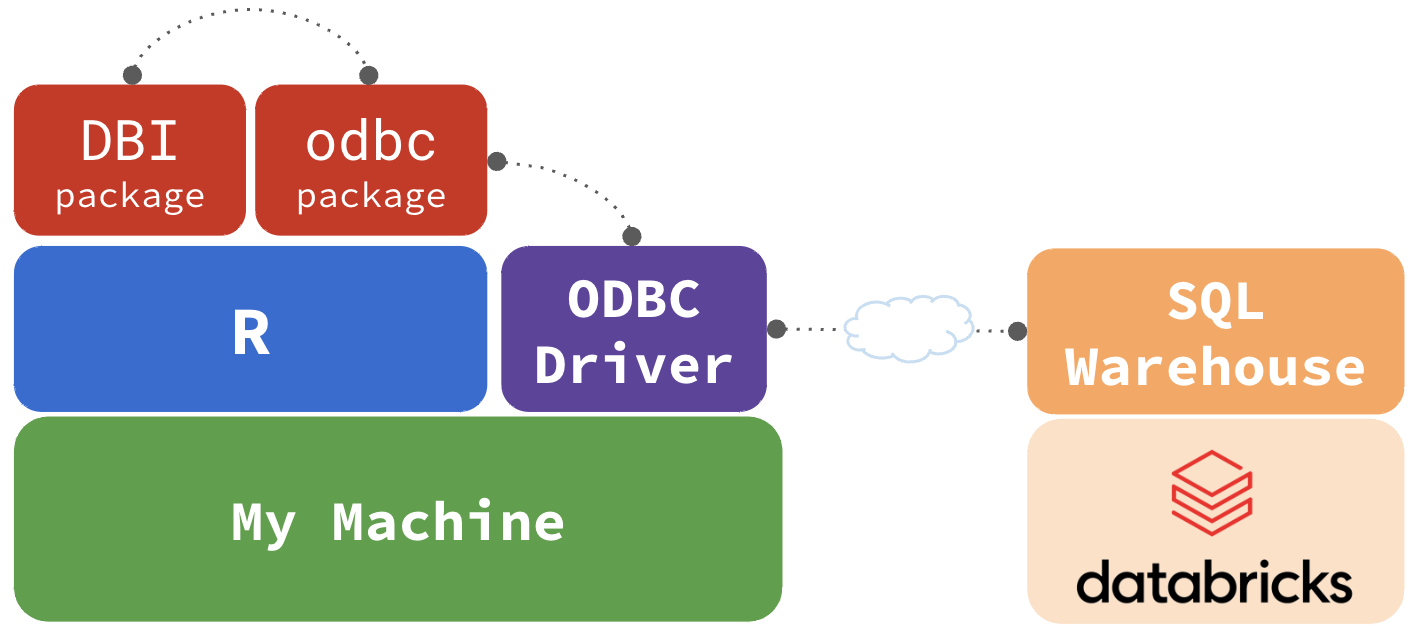
Connecting - Good
Easily connect with dbConnect() & odbc()
https://solutions.posit.co/connections/db/databases/databricks/#using-the-odbcodbc-function
Connecting - Better

odbc::databricks()simplifies the setup- Automatically detects credentials, driver, and sets default arguments
HTTPPathis the only argument that you will need
https://solutions.posit.co/connections/db/databases/databricks/#using-the-new-odbcdatabricks-function
Exercises 1.3 - 1.4
Using DBI
dbConnect()- ConnectsdbDisconnect()- DisconnectsdbListTables()- Lists the tablesdbListFields()- Lists the fields of a tabledbGetQuery()- Runs query & collects results
Exercise 1.5
How everything connects (Revisited)

Other RStudio interfaces
knitr SQL engine
RStudio SQL script

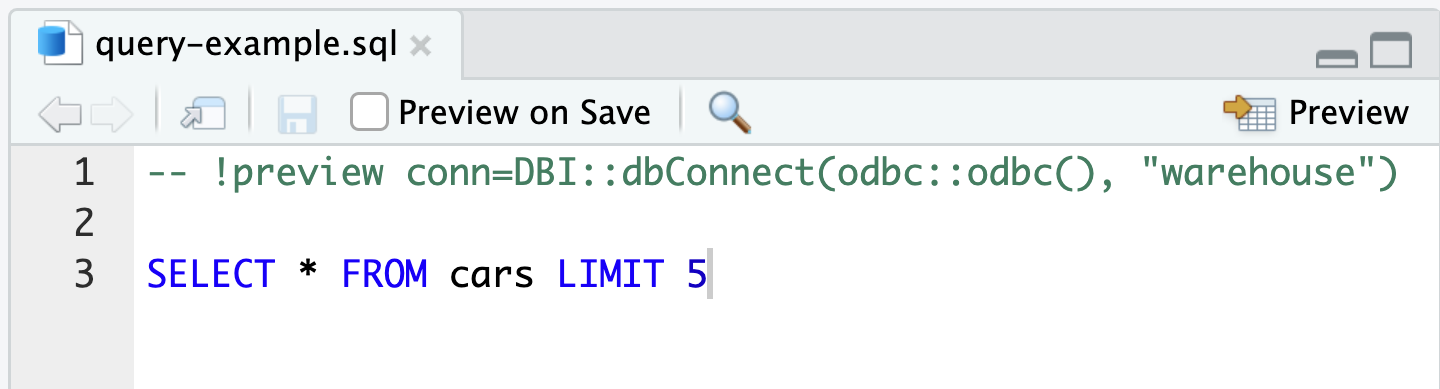
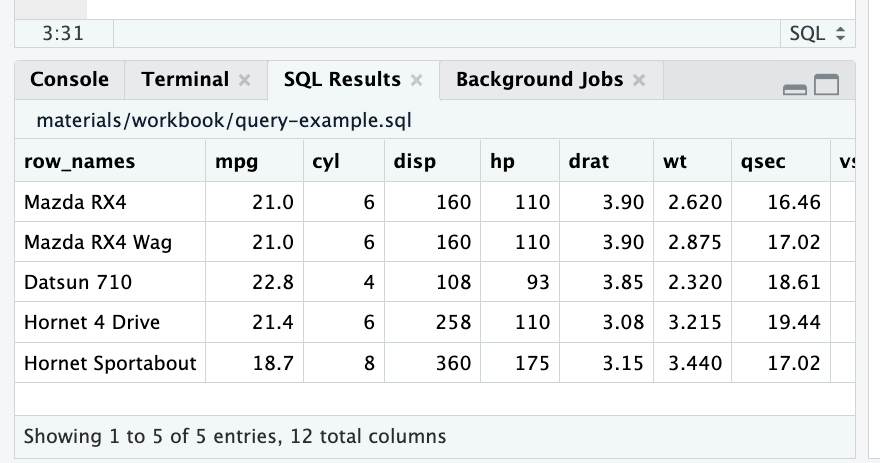
Exercise 1.6 - 1.7
Unit 2
Remote
processing
Approaches
Preferred
“Use in case of emergencies”


SQL not practical
Use dplyr & dbplyr


🔑 How does it work?


🔑 How does it work?


Exercises 2.1 - 2.2
PSA
Please,please,please…don’t pass a full query to tbl()
tbl(con, “Select * from ‘table’ where x = 1”)
PSA
Please, please, please…don’t pass a full query to tbl()
tbl(con, “Select * from ‘table’ where x = 1”)
tbl_table <- tbl(con, “table”)
x_1 <- tbl_table |>
XXfilter(x == 1)
Data collection
There are 3 commands that download data:
print()- Downloads the first 1K rows by default (Guardrails)collect()- Downloads all of the datapull()- Download all the data of a single field
tbl_cars <- tbl(con, “cars”)
print(tbl_cars)
tbl_cars #Print is implied
collect(tbl_cars)
pull(tbl_cars, mpg)
Exercise 2.3
Databricks Unity Catalog (UC)
Centralizes access control, auditing, lineage, and data discovery across workspaces.
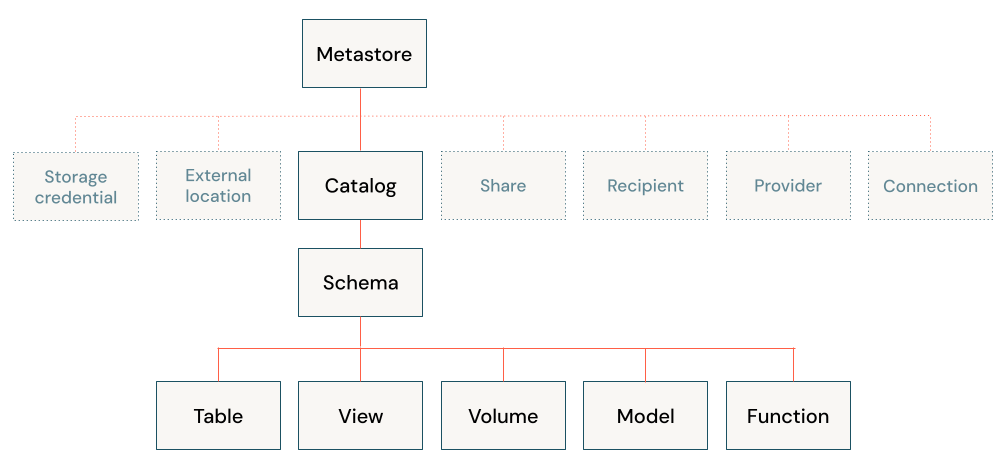
https://docs.databricks.com/en/data-governance/unity-catalog/index.html
Accessing the UC
“diamonds” can be accessed without specifying catalog and schema
# Why do you work?!
tbl(con, “diamonds”)
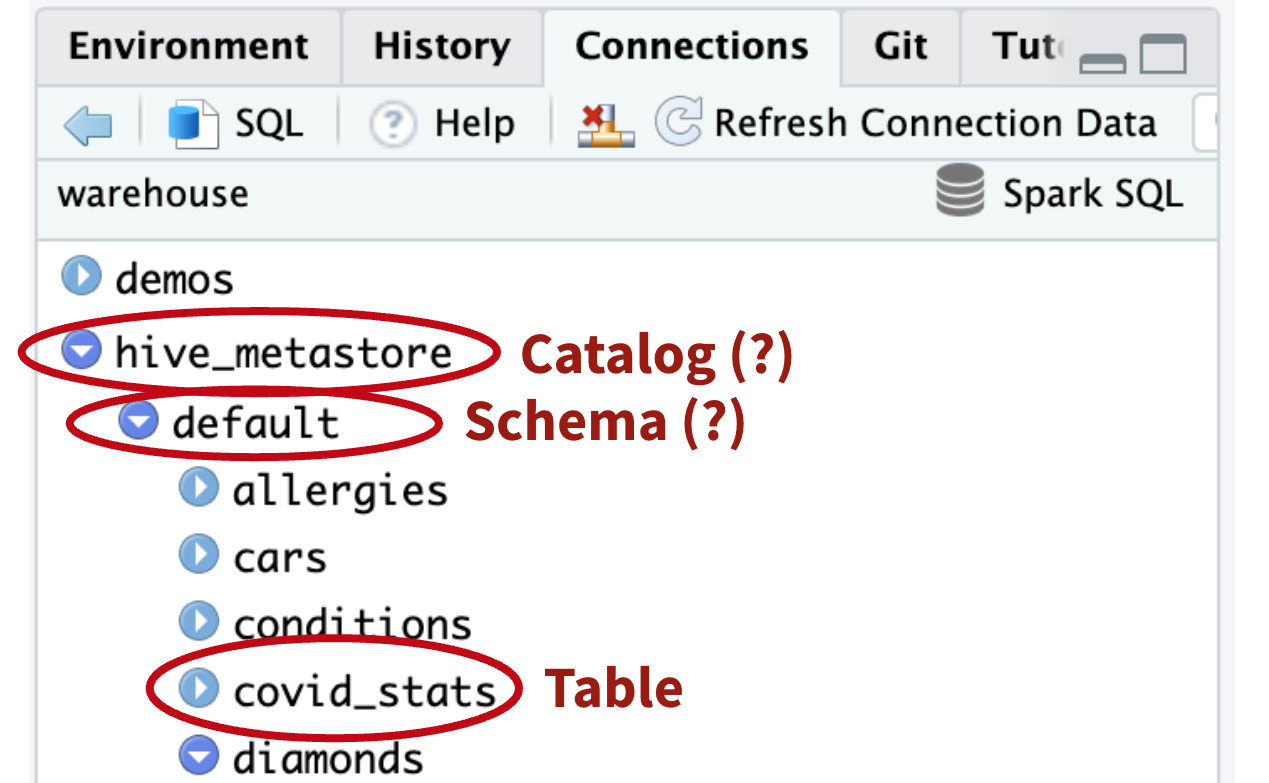
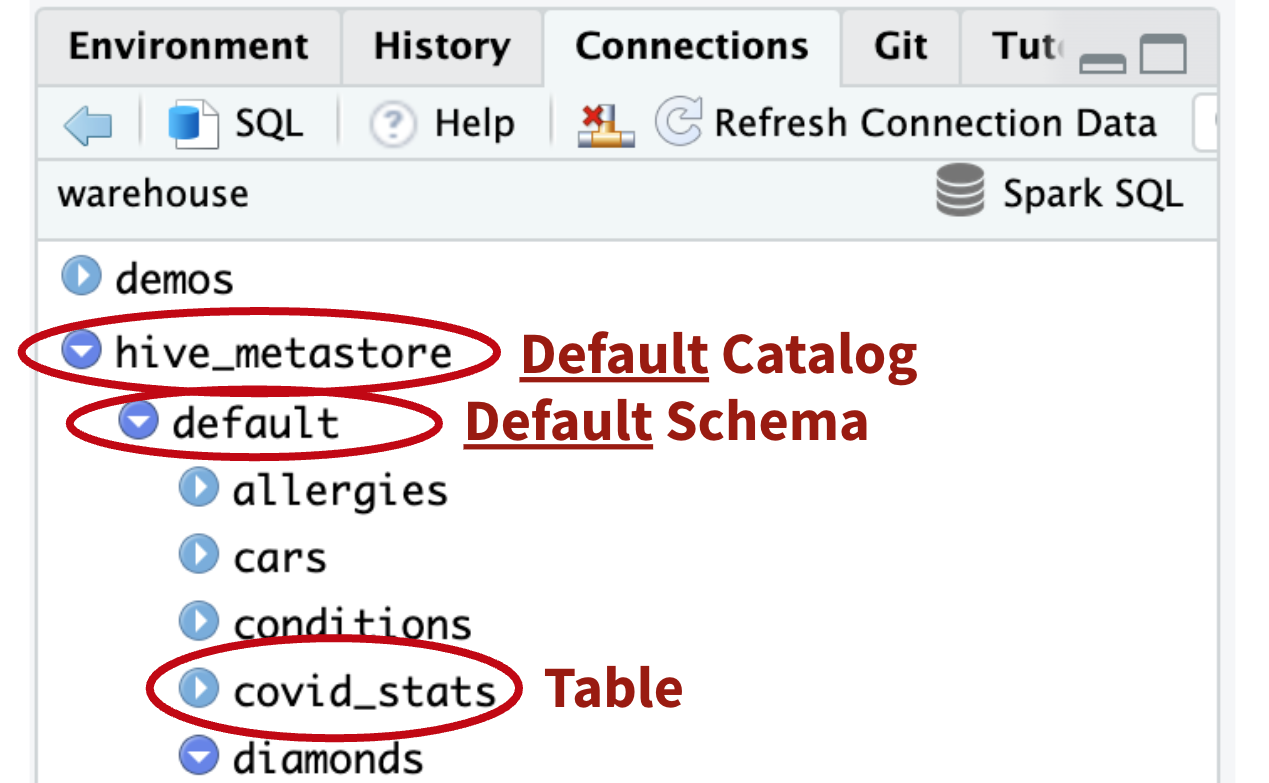
Accessing the UC
“hive_metastore” & “default” are the default catalog & schema respectively
# Oooh, that’s why!
tbl(con, “diamonds”)
Non-default catalog
How do I access tables not in the default catalog?
tbl(con, “customer”)
x (31740) Table or view not found: ..customer
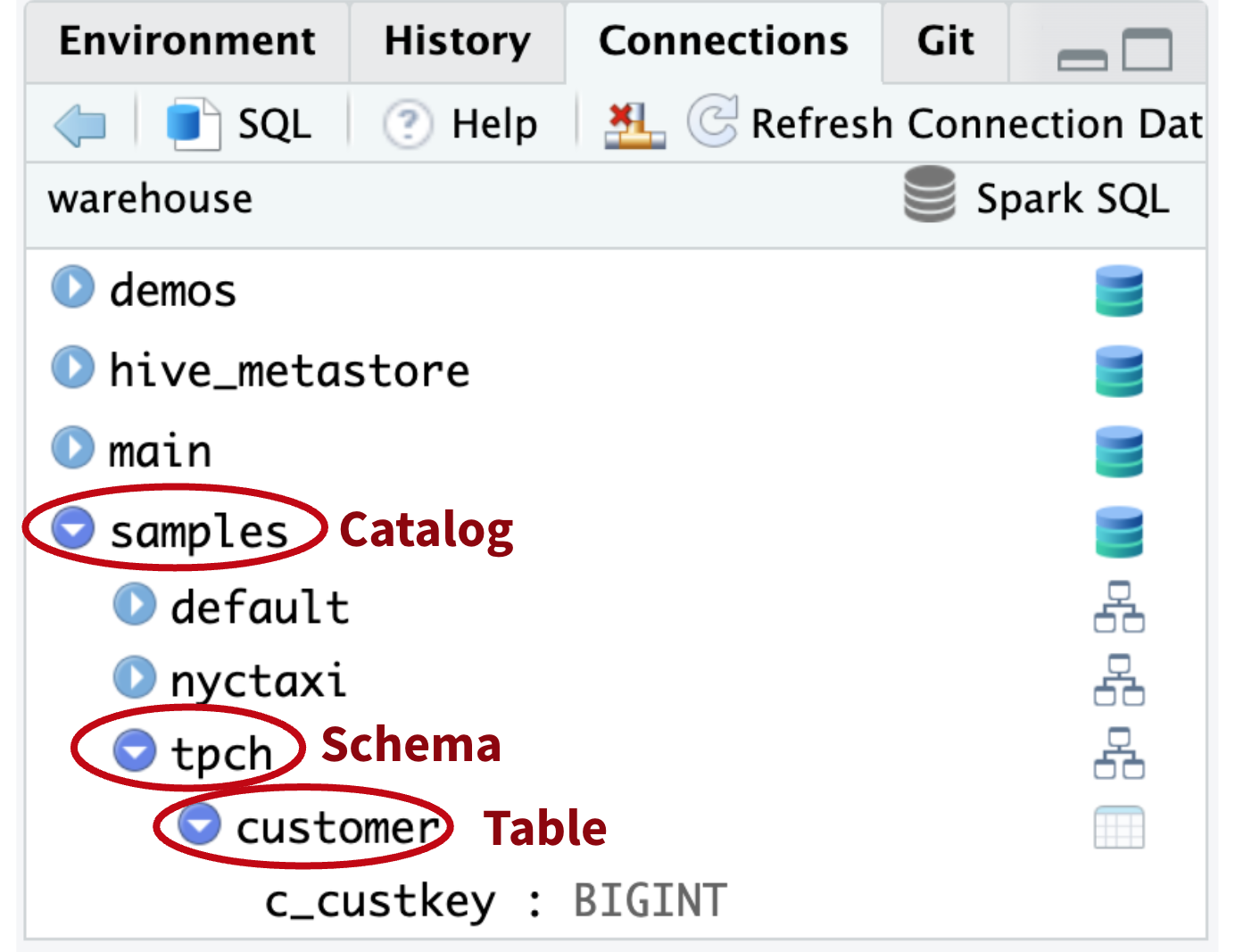
Non-default catalog
How do I access tables not in the default catalog?
tbl(con, “customer”)
x (31740) Table or view not found: ..customer
tbl(
Xcon,
X“workshops.tpch.customer”
X)
x (31740) Table or view not found: ..workshops.tpch.
customer

Non-default catalog
Use I() to create the correct table reference
tbl(
Xcon,
XI(“workshops.tpch.customer”)
X)

https://www.tidyverse.org/blog/2024/04/dbplyr-2-5-0/#referring-to-tables-in-a-schema
Exercise 2.4
Unit 3
Preparing
and exploring
data
Variable selection
Using select() with these functions make variable selection easy:
starts_with()- Starts with an exact prefixends_with()- Ends with an exact suffixcontains()- Contains a literal stringeverything()- Selects everything else. Use with other selection functions.
Exercise 3.1
Joining tables

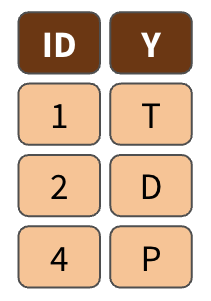
left_join()
right_join()
full_join()
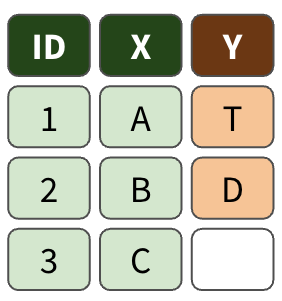
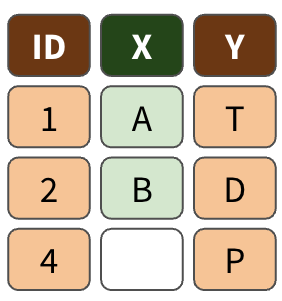

Exercise 3.2
Preparing data
Load prepared data to a variable, without downloading it
prep_orders <- orders |>
left_join(customers, by = c("o_custkey" = "c_custkey")) |>
left_join(nation, by = c("c_nationkey" = "n_nationkey")) |>
mutate(
order_year = year(o_orderdate),
order_month = month(o_orderdate)
) |>
rename(customer = o_custkey) |>
select(-ends_with("comment"), -ends_with("key"))Exercise 3.3
Exploring data
Use the base query for data exploration
Exercise 3.4
Unit 4
Visualizations
Plotting local data
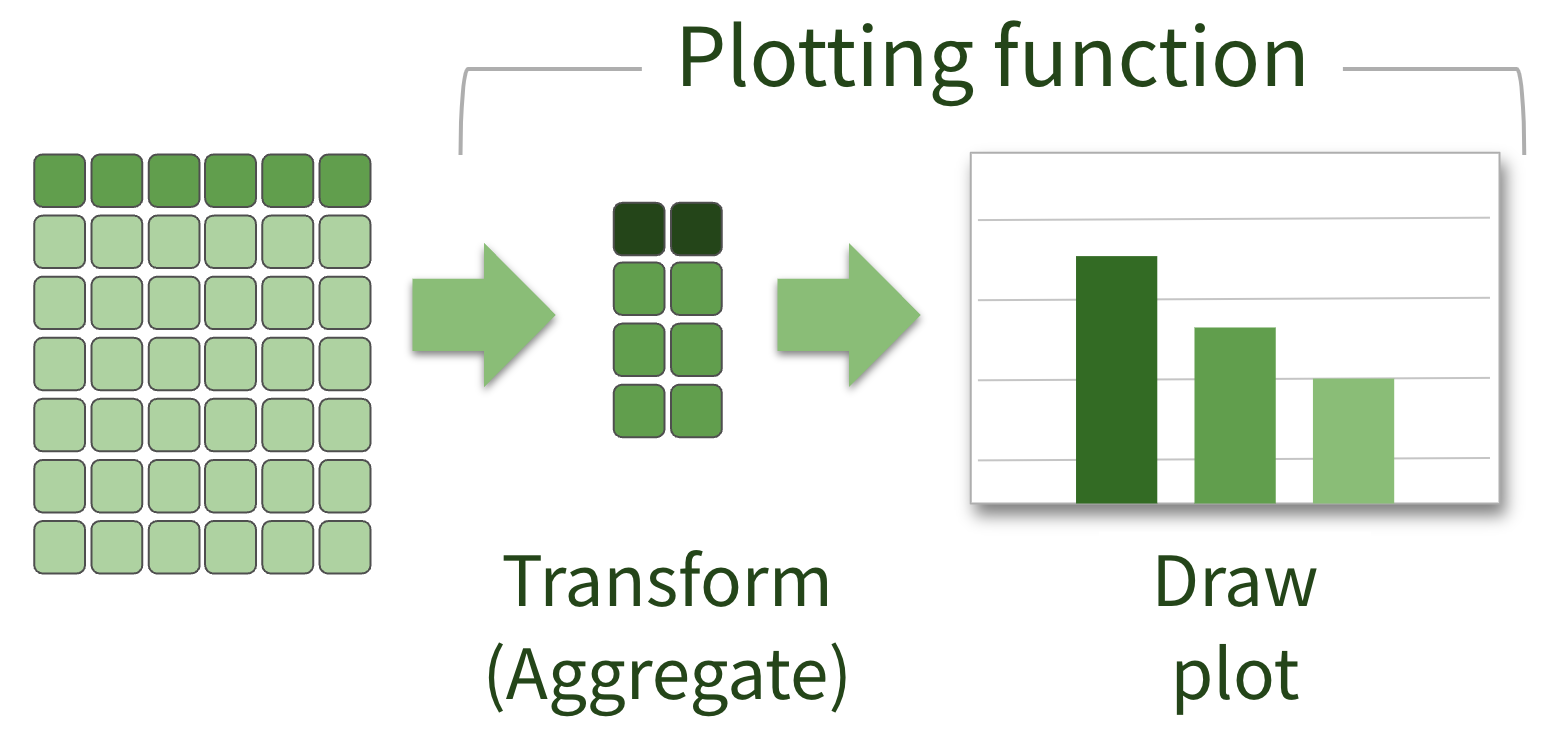
Plotting remote data
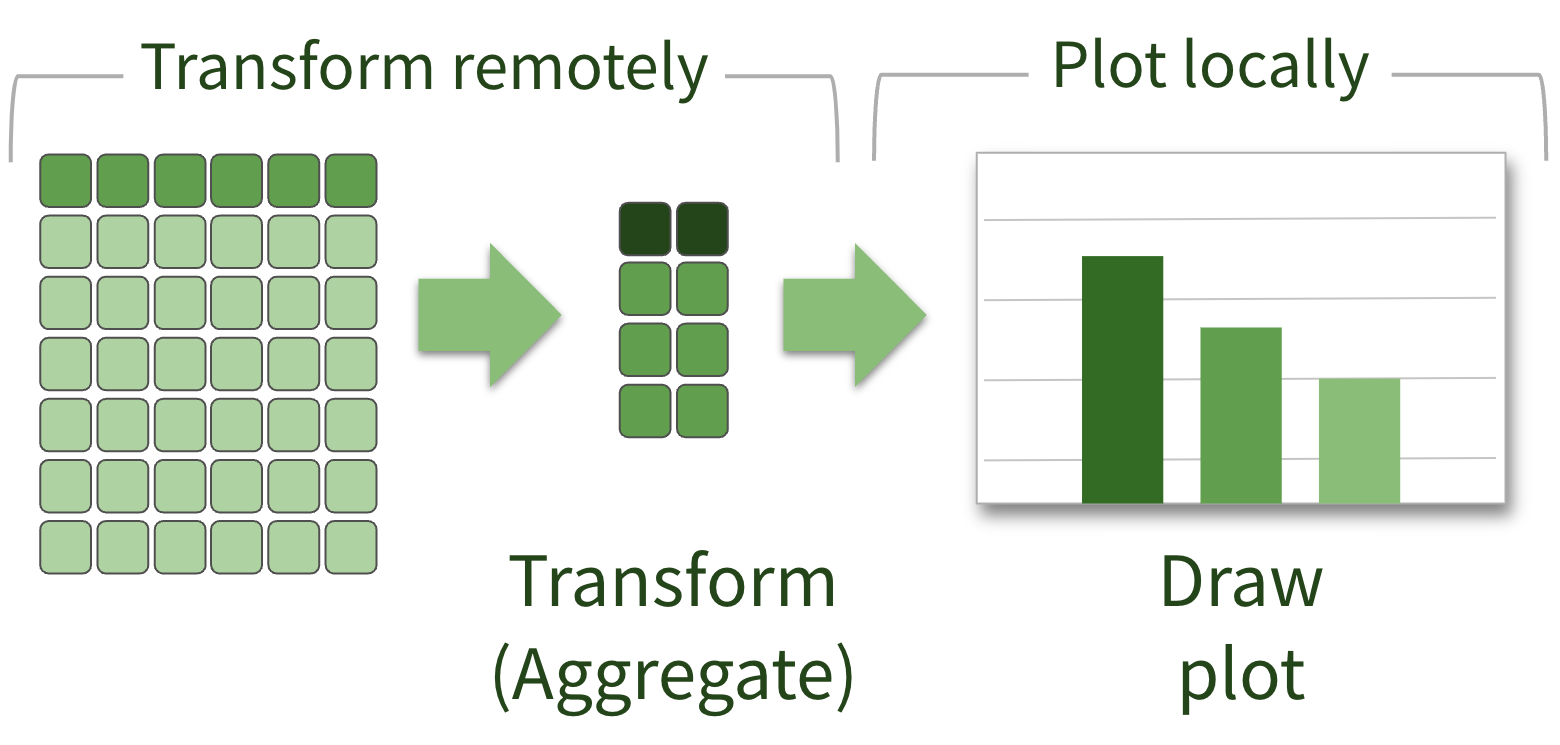
What that looks like in R
Pipe the prepared data into a ggplot2

What that looks like in R
Pipe the prepared data into a ggplot2
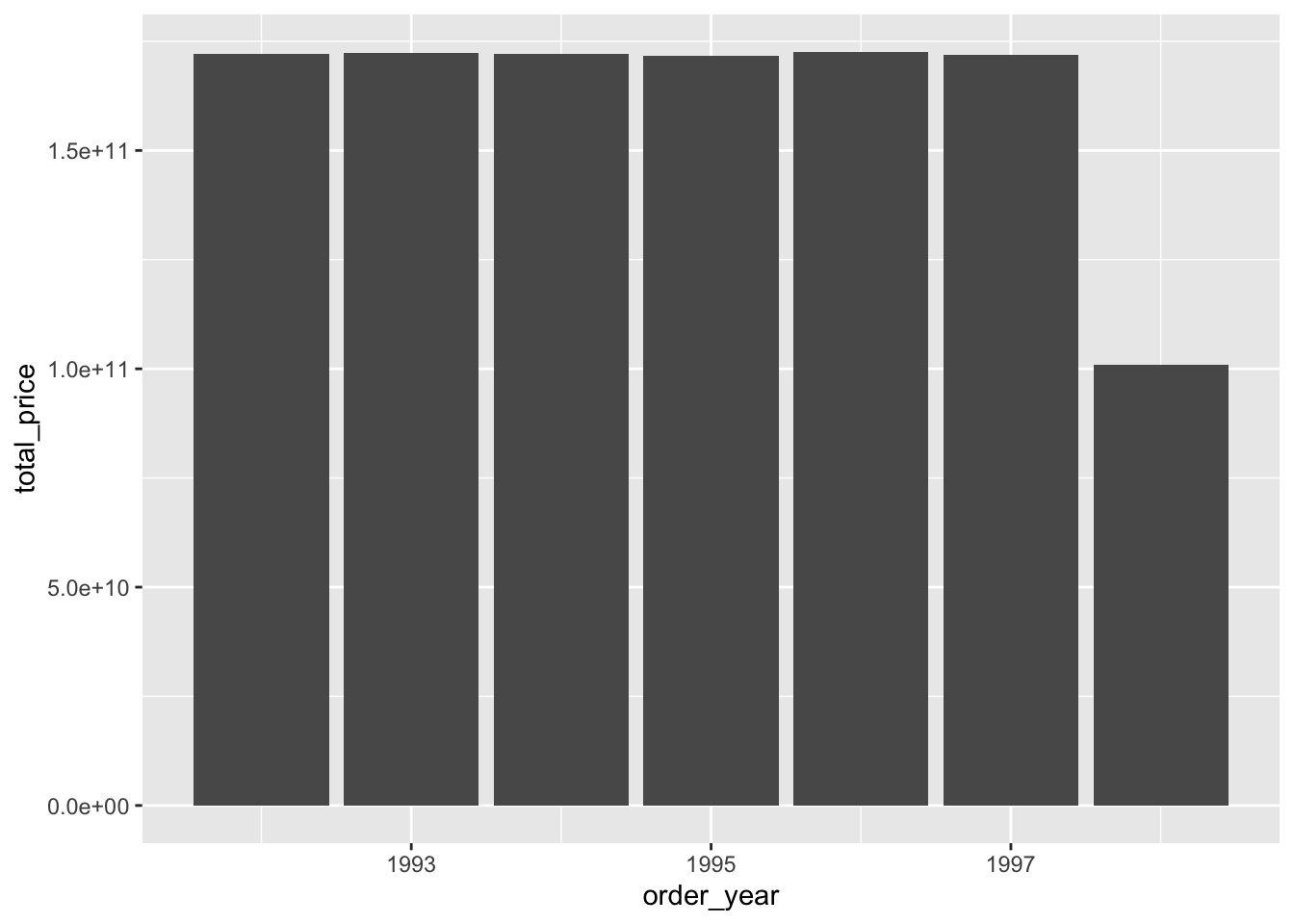


What that looks like in R
Pipe the prepared data into a ggplot2

ggplot2 “auto-collects”
Be careful! It downloads all of the data!

Exercise 4.1
The truth about ggplot2
A plot gets refined iteratively, data must be local.
- ‘Improve scales’
- ‘Add labels to the data’
- ‘Add title and subtitle’
- ‘Improve colors’
- ‘Change the theme’
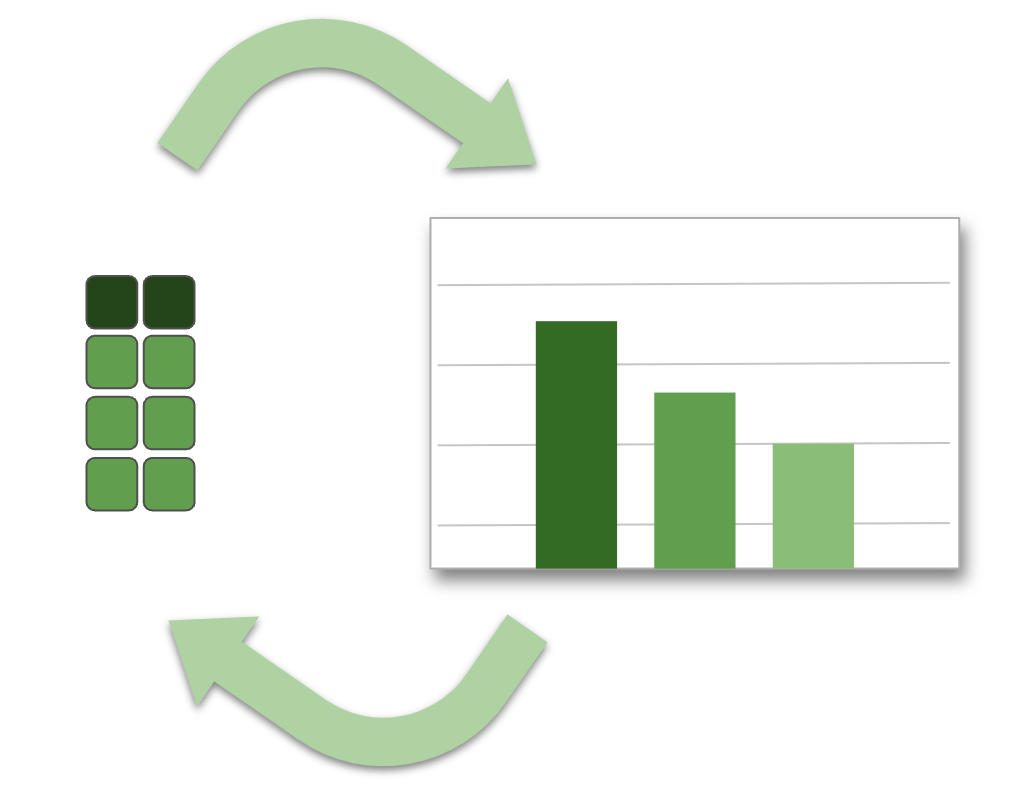
What that looks like in R
Collect (download) the data into an R variable
What that looks like in R
Iterate on the plot using the local data
What that looks like in R
Iterate on the plot using the local data
breaks <- as.double(quantile(c(0, max(sales_by_year$total_price))))
breaks_labels <- paste(round(breaks / 1000000000, 1), "B")
sales_by_year |>
ggplot() +
geom_col(aes(order_year, total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year") +
theme_light()Exercises 4.2 - 4.4
Unit 5
Databricks
Connect
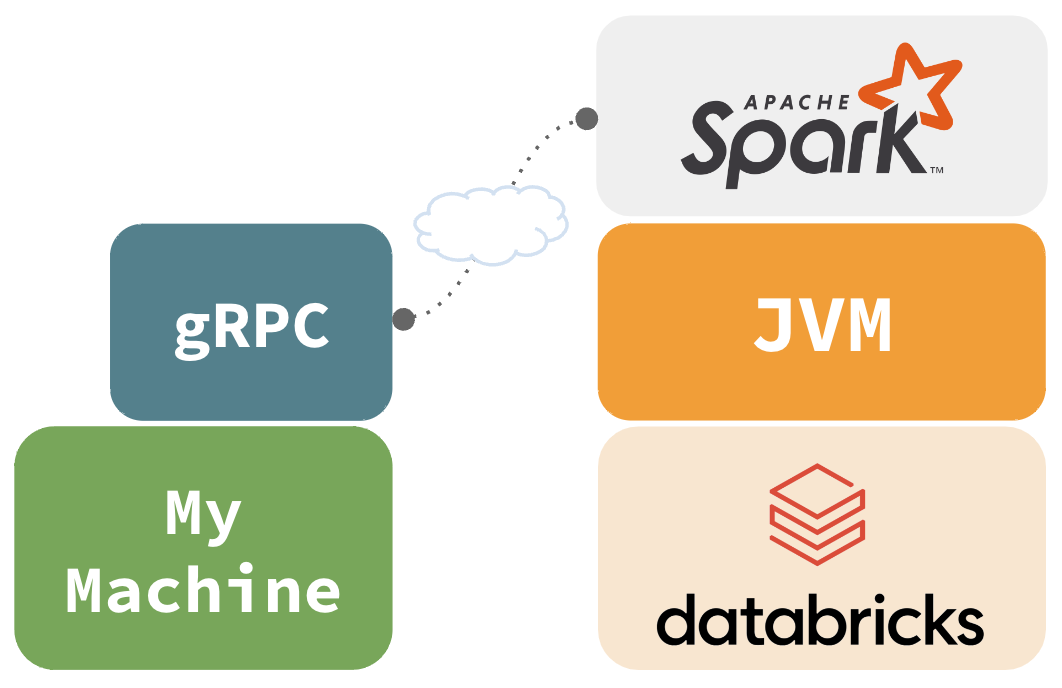
Databricks Connect
- Spark Connect, offers true remote connectivity
- Uses gRPC to as the communication interface
- Databricks Connect ‘v2’ is based on Spark Connect (DBR 13+)
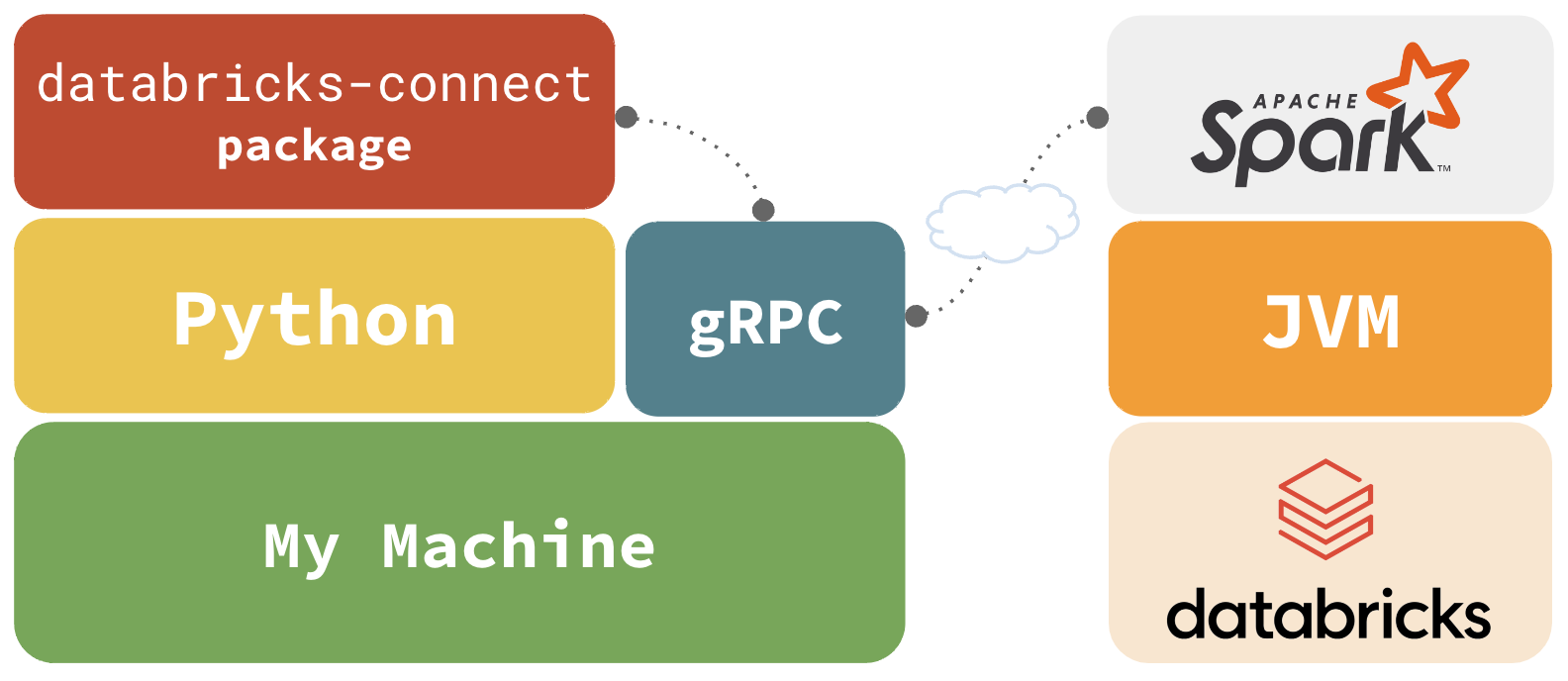
Databricks Connect
databricks-connect integrates with gRPC via pyspark
Databricks Connect

sparklyr integrates with databricks-connect via reticulate
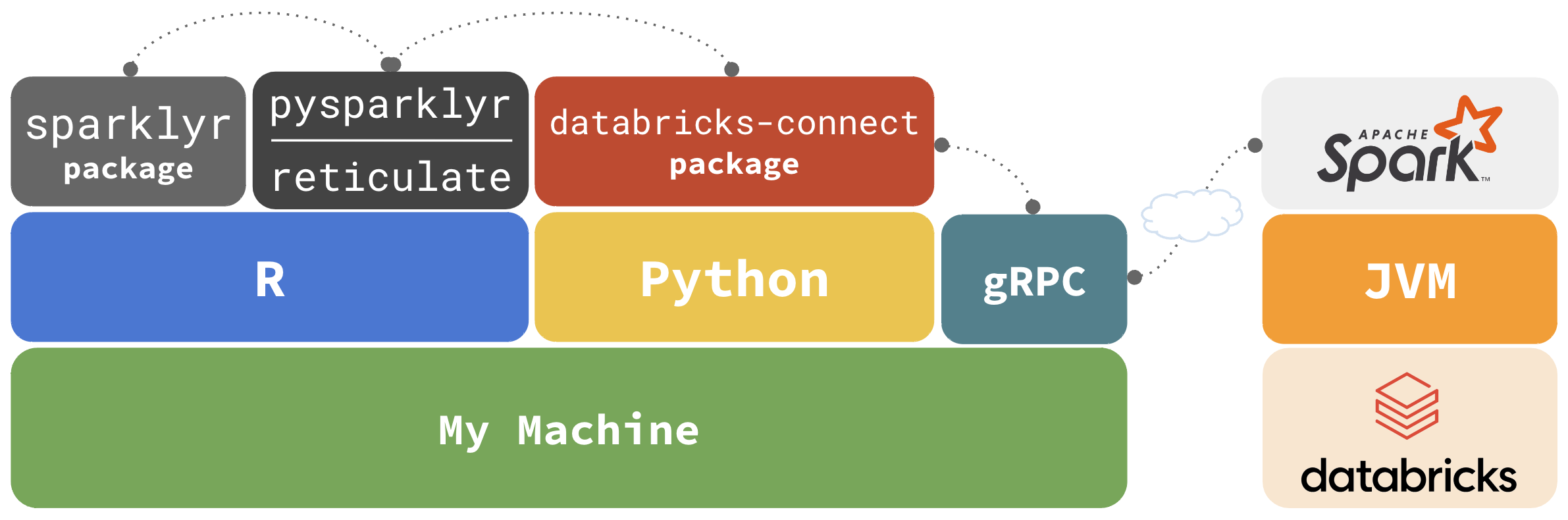
Why not just use ‘reticulate’?
sparklyr extends functionality and user experience
dplyrback-endDBIback-end- R UDFs
- RStudio, and Positron, Connections Pane integration

Getting started

Getting started

Automatically, checks for, and installs the Python packages
install.packages("pysparklyr")
library(sparklyr)
sc <- spark_connect(
cluster_id = "[cluster's id]",
method = "databricks_connect"
)
#> ! Retrieving version from cluster '1026-175310-7cpsh3g8'
#> Cluster version: '14.1'
#> ! No viable Python Environment was identified for
#> Databricks Connect version 14.1
#> Do you wish to install Databricks Connect version 14.1?
#> 1: Yes
#> 2: No
#> 3: CancelExercise 5.1
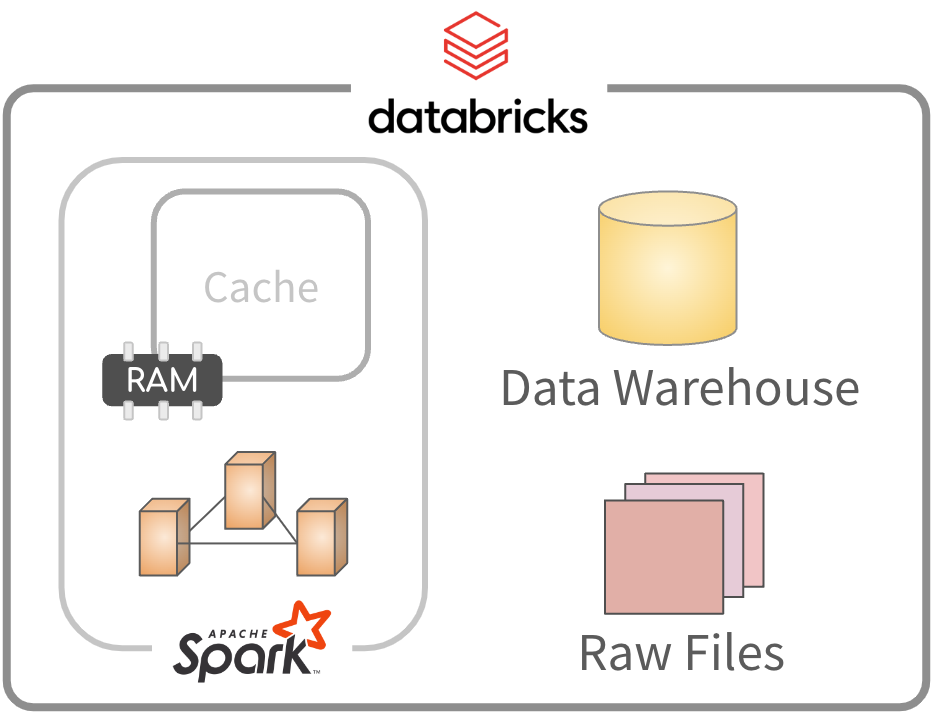
Understanding Spark’s data caching
Spark’s data capabilities
- Spark has the ability to cache large amounts of data
- Amount of data is limited by the size of the cluster
- Data in cache is the fastest way to access data in Spark
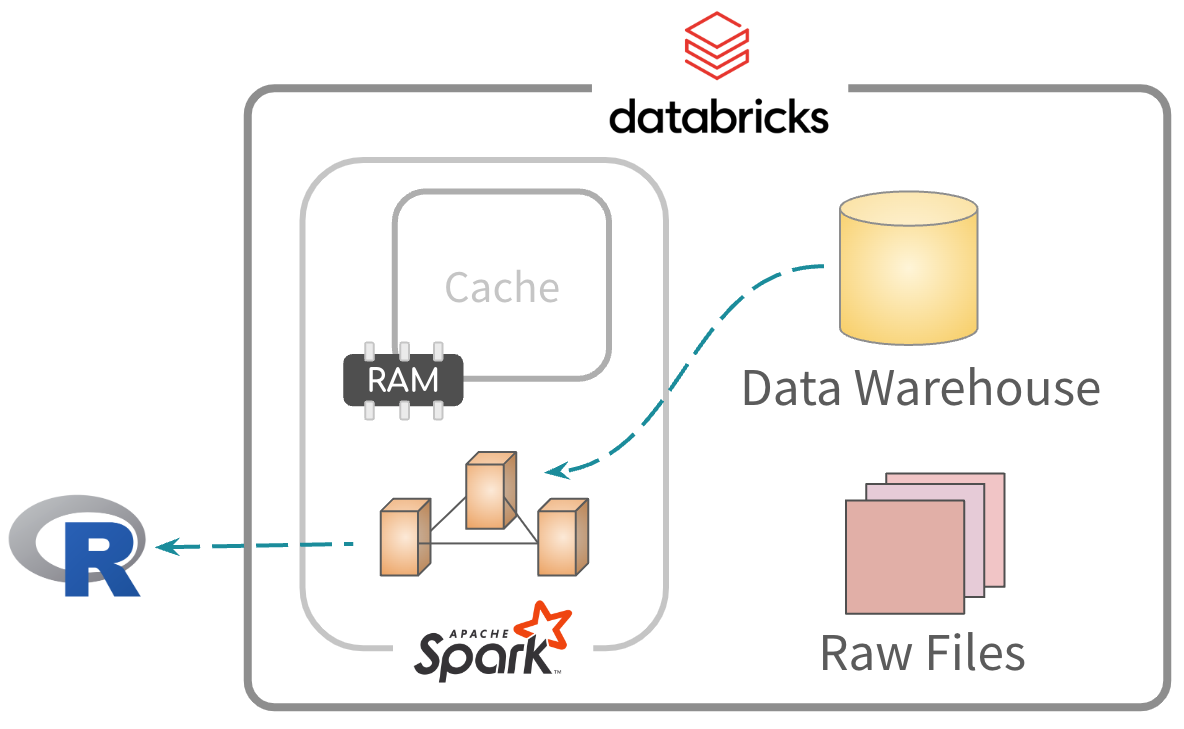
Default approach
Data is read and processed. Results go to R.
About this approach
- Well suited when exploring the entirety of the data. Usually to find relevant variables
- Not efficient when accessing the same fields and rows over and over
Uploading data from R
copy_to() to upload data to Spark
Exercise 5.2
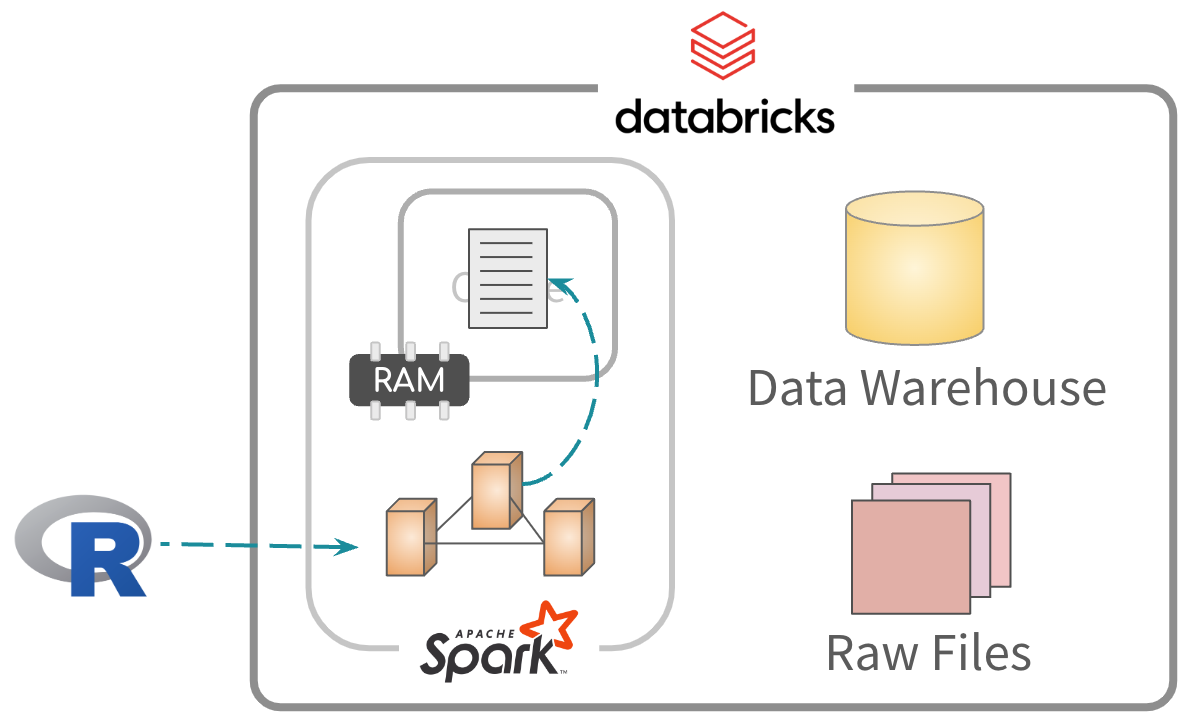
Caching data
2 step process. first, cache all or some data in memory
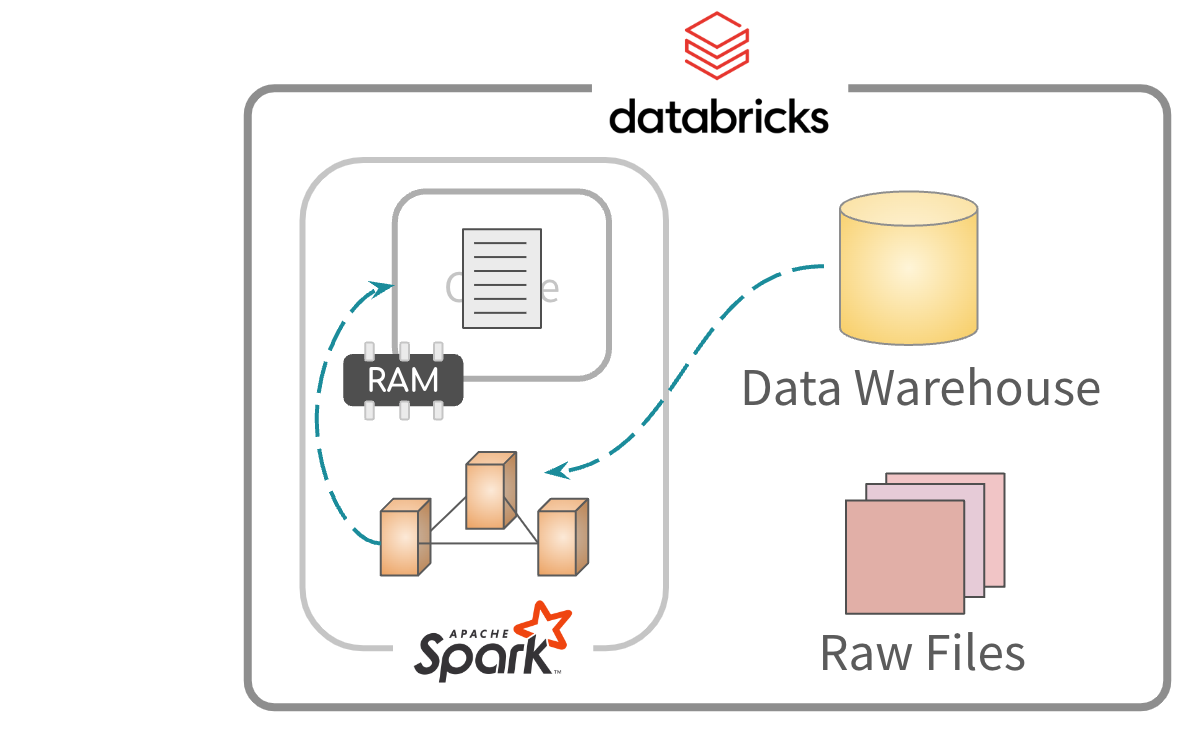
Caching data
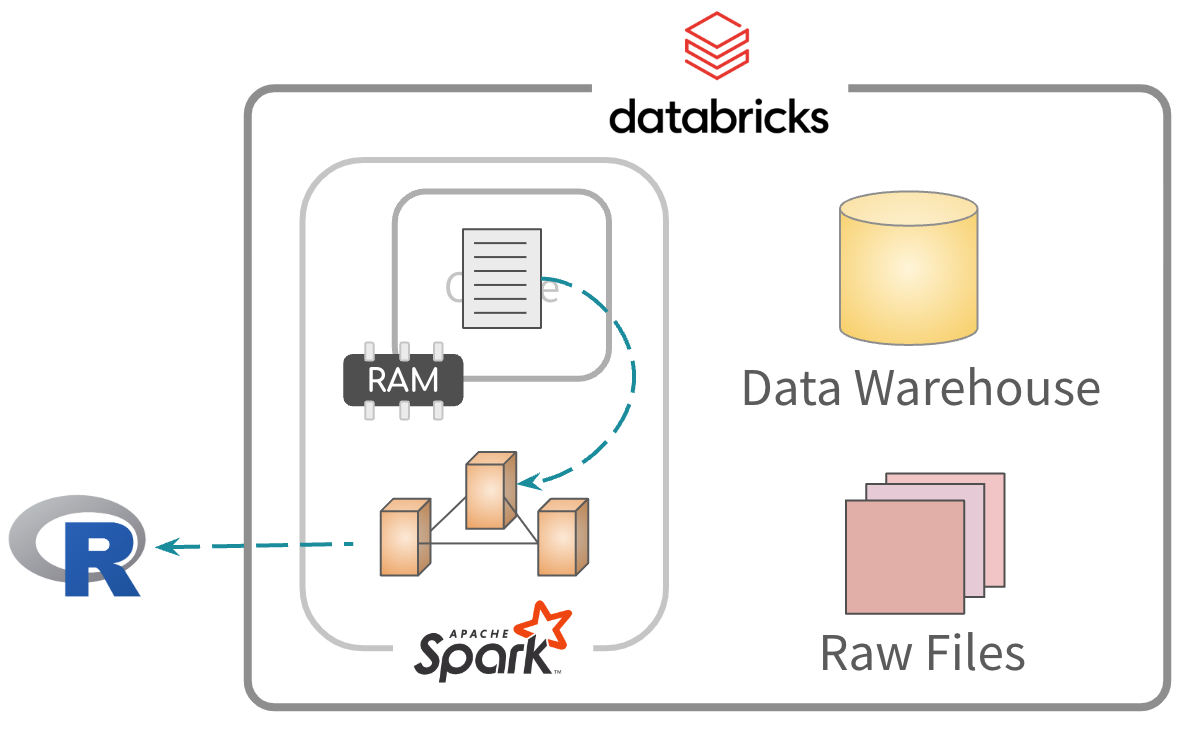
Second, read and process from memory. Much faster
About this approach
- Well suited when accessing the same fields and rows over and over
- Best when running models and UDFs (More about this later)
Exercise 5.3
Reading files
By default, files are read and saved to memory
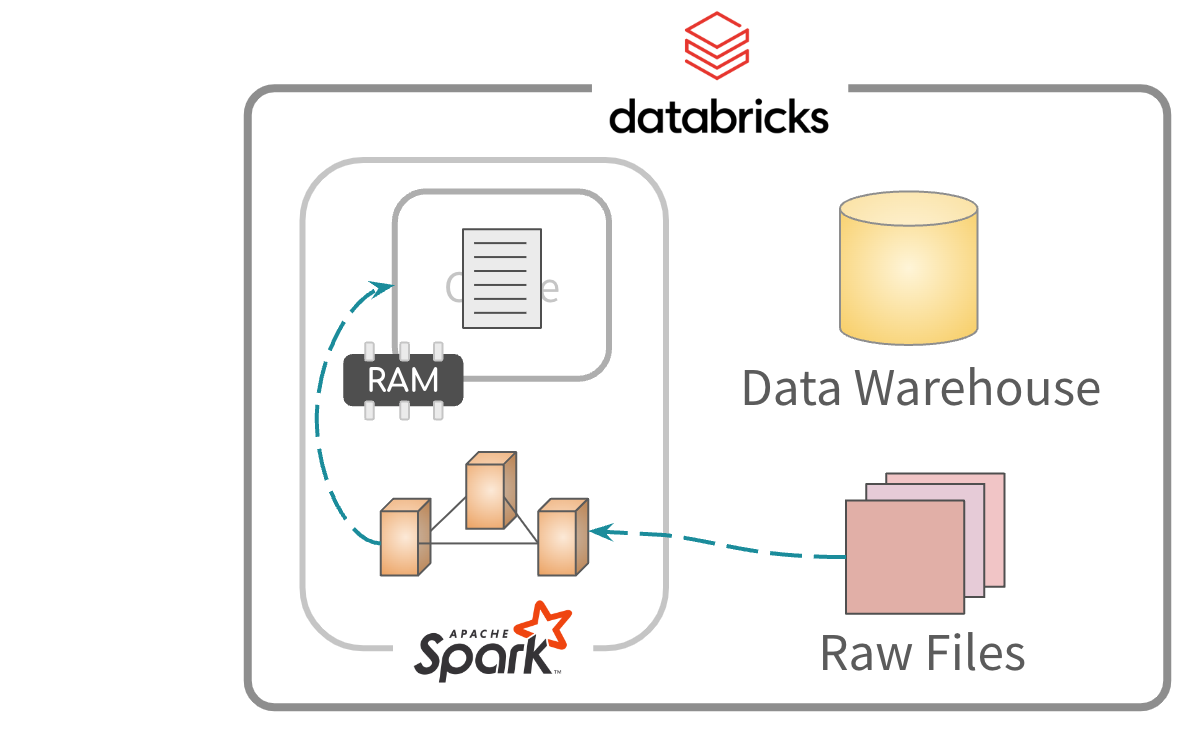
Reading files
Afterwards, the data is read from memory for processing

About this approach
- Read files using the
spark_read...family of functions - The file path needs to be relative to your Databricks environment
- Databricks Volumes are ideal for this approach
Exercise 5.4
“Mapping” files
The files can be mapped but not imported to memory
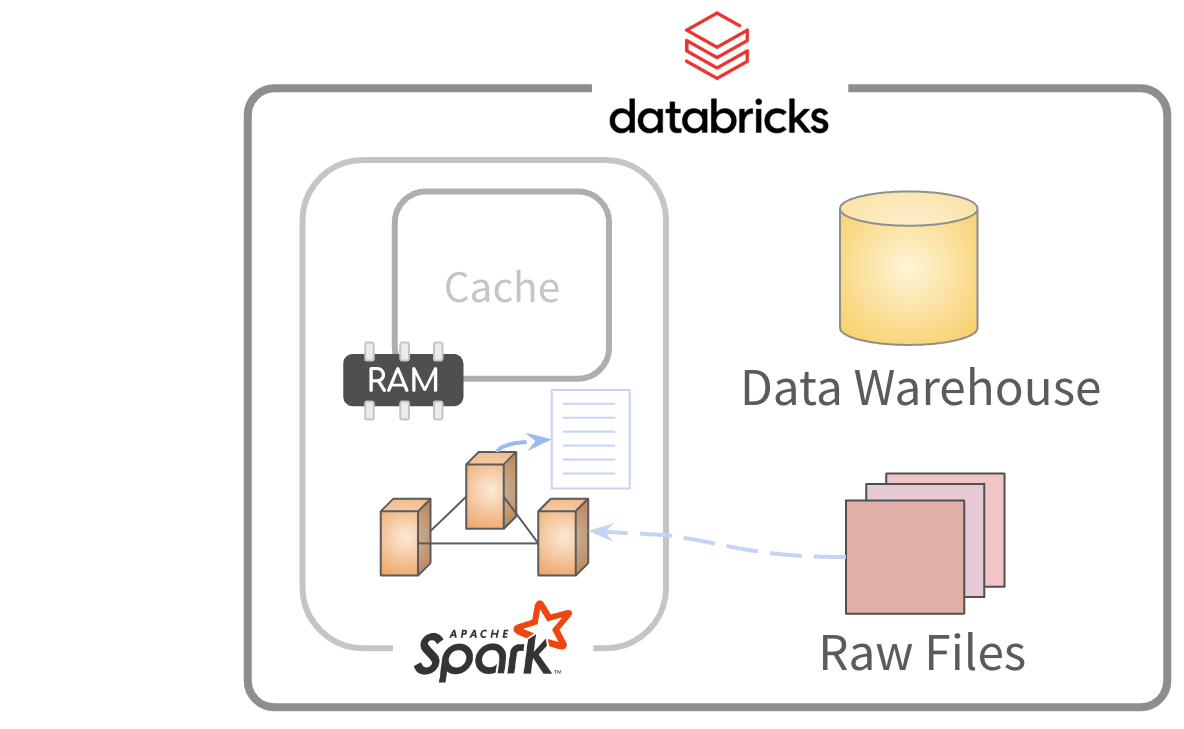
“Mapping” files
Data is read and processed. Results sent to R.
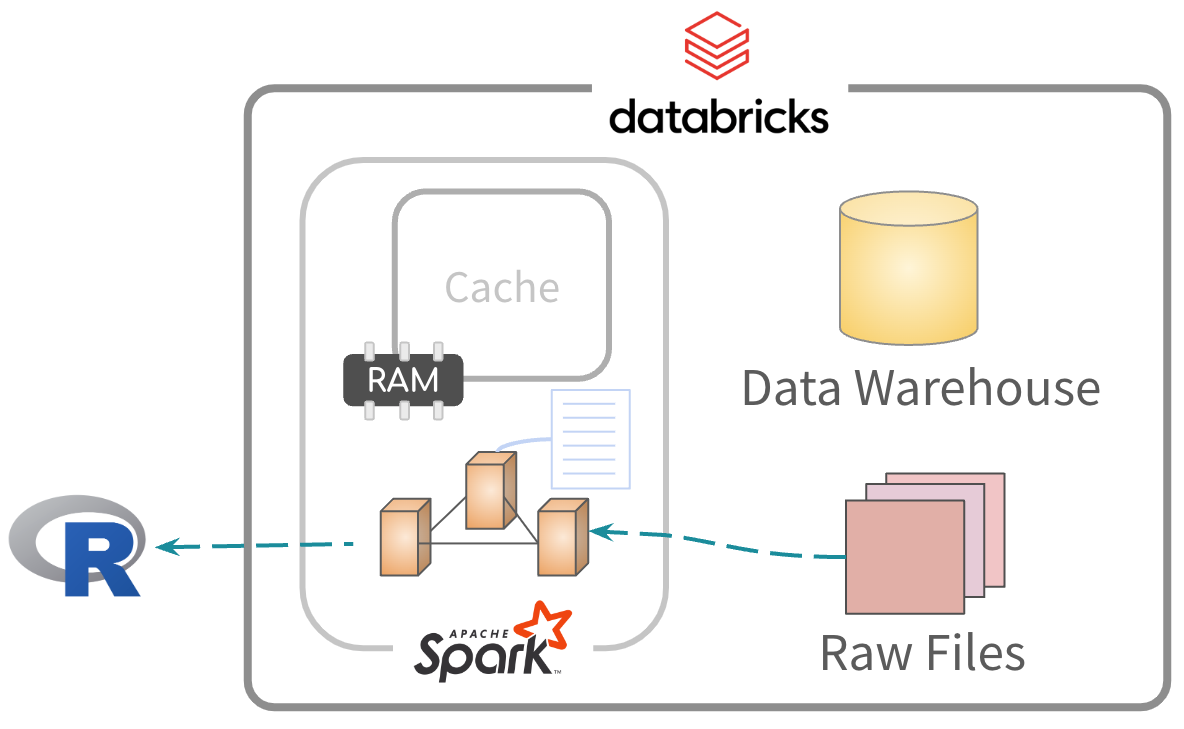
Exercise 5.5
Partial cache
Alternatively, you can cache specific data from the files
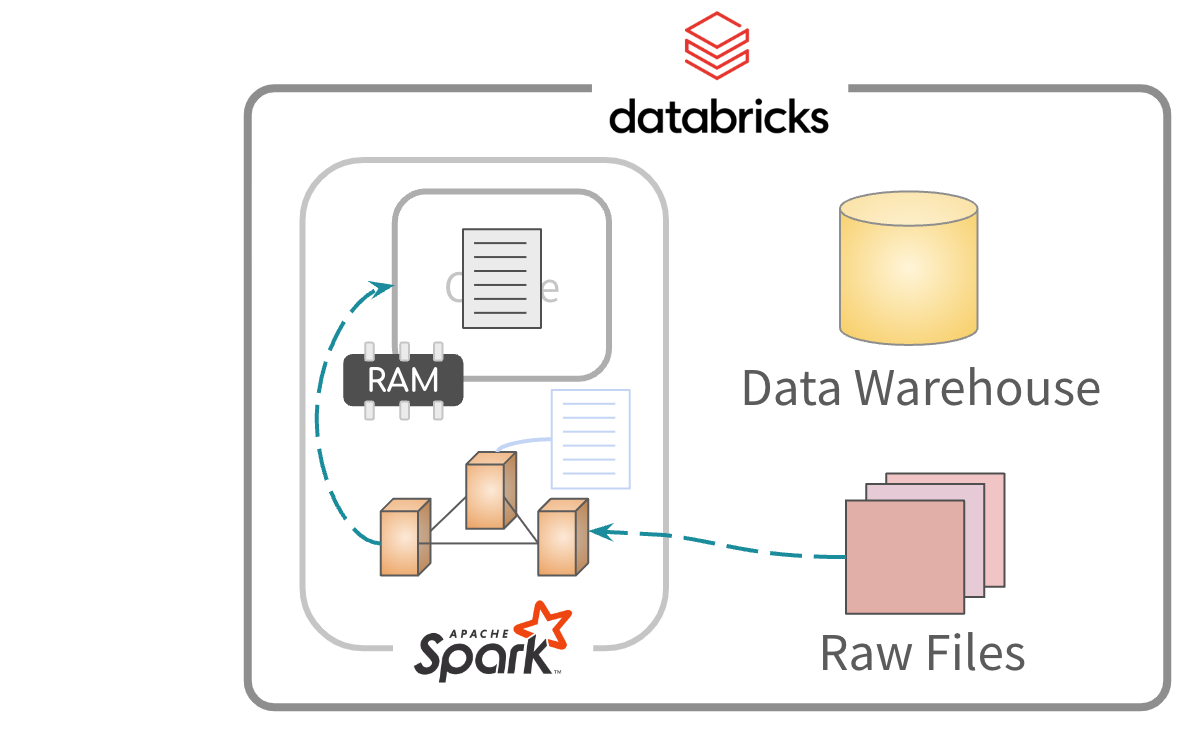
Partial cache
Afterwards, the data is read from memory for processing

Exercise 5.6
Very large files, read or map
- Reading, you “pay” in time at the beginning
- Mapping, you “pay” in time as you access the data
- Extended EDA, reading would be better
- EDA of targeted data, partial caching would be better
- Jobs that pull a predetermined set of data, mapping would be better
End game
Combine the data from any approach. Cache the resulting table
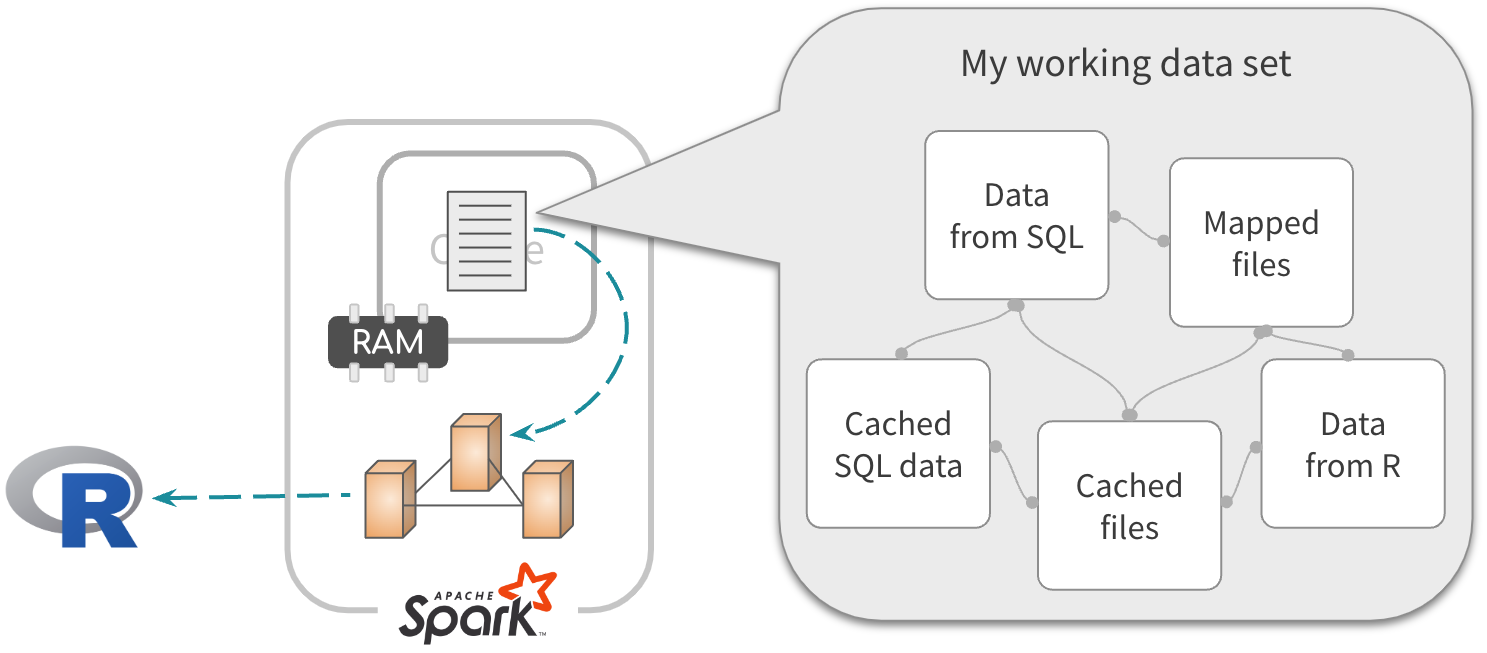
Exercise 5.7
Unit 6
Intro to
R UDFs
What is an UDF?
- Stands for “User Defined Function”
- Enables operations not built-in Spark
- Can be written in Scala, Python, or R
Ok, but what does that mean?!
Ok, but what does that mean?!
We can run R code inside Spark! 🎉 🎉
But first… parallel processing
Spark partitions the data logically


Parallel processing
Each node gets one, or several partitions








Parallel processing
The cluster runs jobs that process each partition in parallel












Parallel processing
Your R function runs on each partition independently















Parallel processing
Results are appended, and returned to user as a single table




Accessing in R

Exercise 6.1
Group by variable

Use group_by to override the partitions, and divide data by a column
Exercise 6.2
Custom functions

R function should expect and return a table

Exercise 6.3
R packages

R packages have to be referenced from within your function’s code using library() or ::
tbl_mtcars |>
spark_apply(function(x) {
library(broom)
model <- lm(mpg ~ ., x)
tidy(model)[1,]
},
group_by = "am"
)
#> # Source: table<`sparklyr_tmp`> [2 x 6]
#> # Database: spark_connection
#> am term estimate std_error statistic p_value
#> <dbl> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 0 (Intercept) 8.64 21.5 0.402 0.697
#> 2 1 (Intercept) -138. 69.1 -1.99 0.140Exercise 6.4
Dependencies in Databricks
Spark clusters are tied to a Databricks Runtime version. Each version of DBR contains specific versions of:
- Spark
- Java
- Scala
- Python
- R
- Set of Python and R packages
https://docs.databricks.com/en/release-notes/runtime/15.3.html#installed-r-libraries
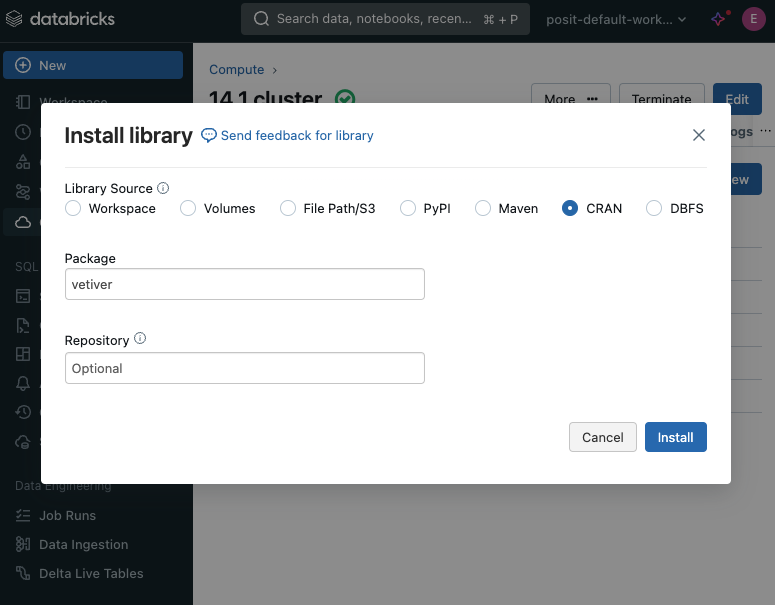
Missing packages
- Additional packages can be installed
- In the Databricks portal, use the Libraries tab of the cluster
- Python packages will install from PyPi
- R packages will install from CRAN
Key considerations
- You are dealing with two different environments
- Your Python version must match the cluster
- R version can be different, but could be source of errors
- Same for Python, and R packages. OK if mismatch, but could be errors
Key considerations
An example of different R versions
Exercise 6.5
Solving for R UDFs
Solving for R UDFs
It wasn’t easy…
Problem
Spark Connect does not support R UDFs natively
Problem
Spark Connect does not support R UDFs natively
(Databricks Connect v2, is based on Spark Connect)
In Python
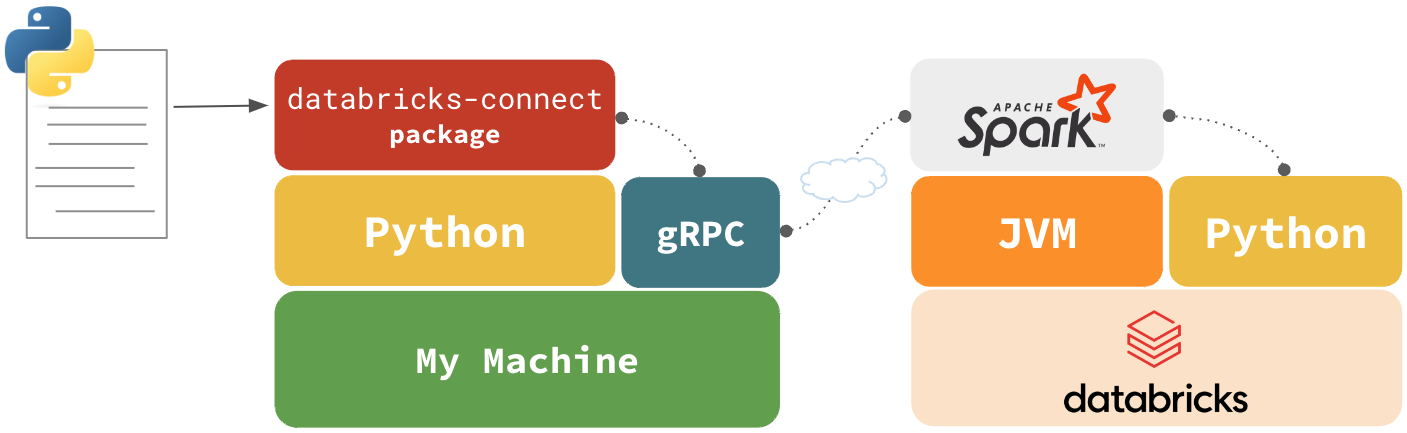
Code sent through gRPC, code runs in remote Python environment through Spark
R UDFs

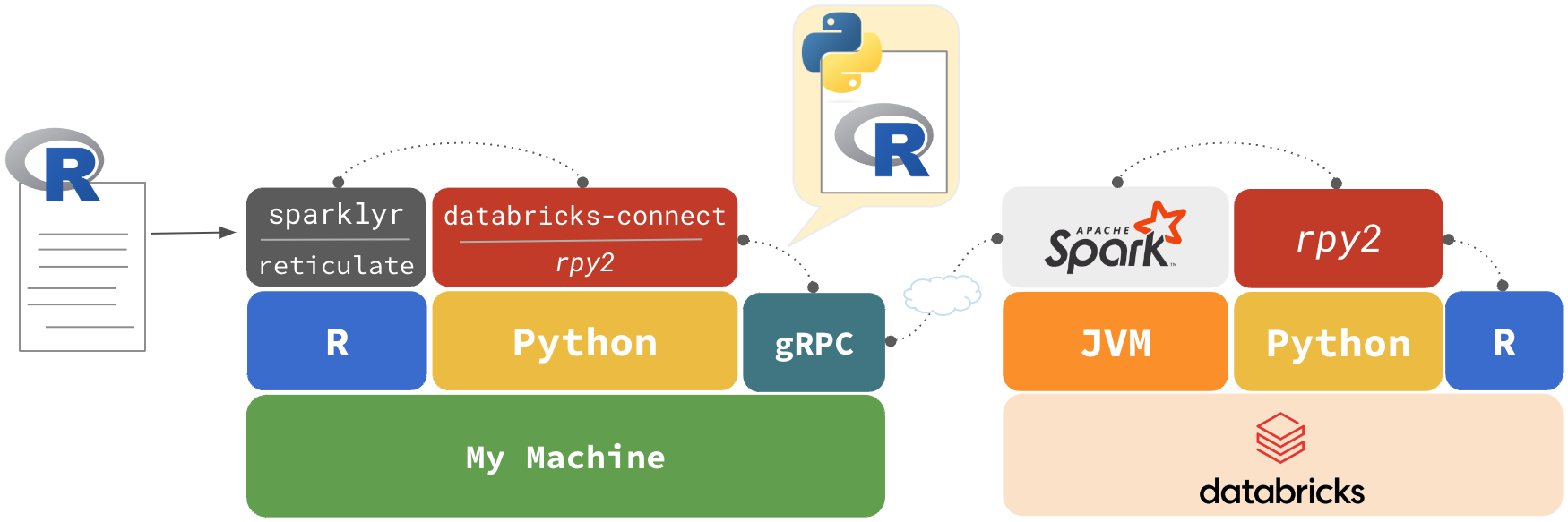
R code is inserted in a Python script, rpy2 executes
https://spark.posit.co/deployment/databricks-connect-udfs.html
Wrapped Python functions

R:
Python:
Unit 7
Modeling
with R UDFs
Run R models in Spark
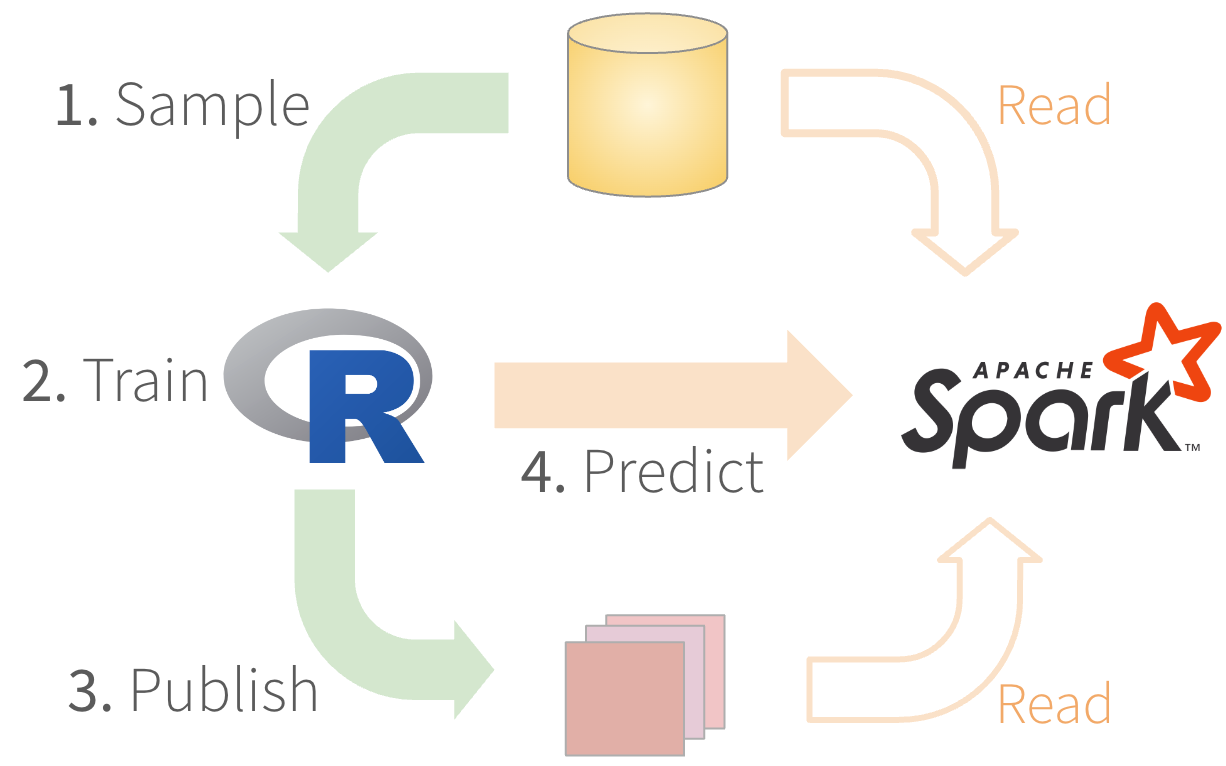
1. Sample - Use dplyr


Exercise 7.1
2. Train - Ideally, tidymodels

Fit the model locally in R
Exercise 7.2
3. Publish - Package with vetiver

Exercise 7.3
3. Publish - Transfer model file
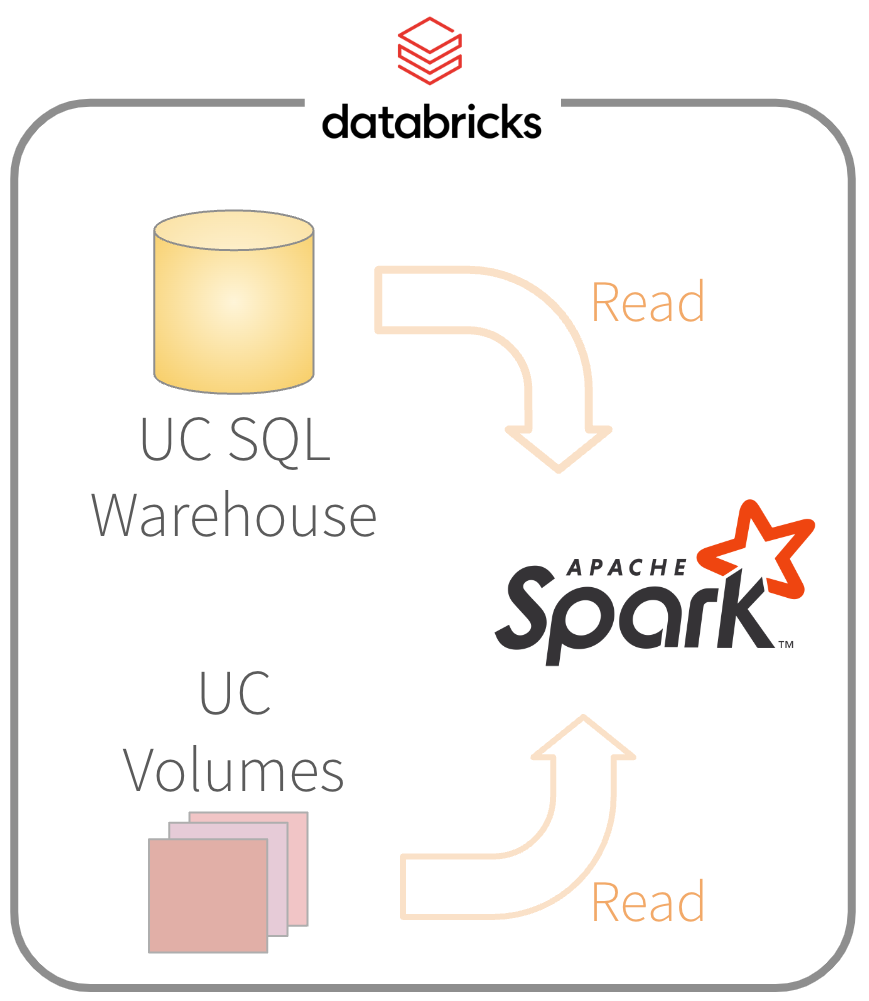
- Copy the R model where Spark can access
- Location has to be secure
- Options: Databricks UC, s3 buckets, Posit Connect, others
3. Publish - Databricks UC Volume
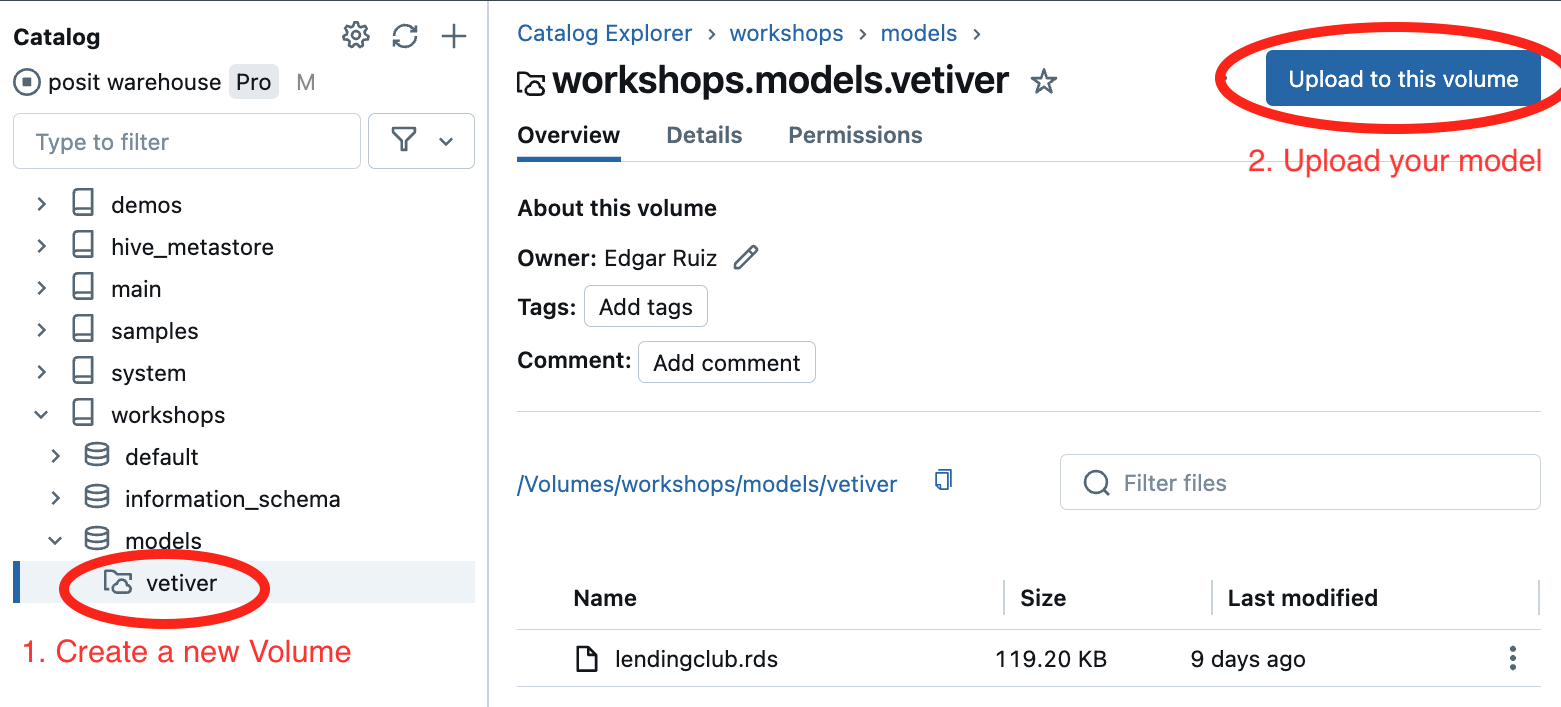
4. Predict - Create the function
It needs to read the model file, runs prediction, and process results
4. Predict - Test the function locally
Good practice to test the function with local data
Exercise 7.4
4. Predict - Switch to published
Function to use the copy of the model in Databricks
4. Predict - Test remotely
Use a sample or top rows to test the function in Spark
4. Predict - Run predictions
Run full set, make sure results are saved
Exercise 7.5
4. Predict
- Create the function
- Test the function locally
- Switch to published model
- Test function remotely
- Run predictions
Run R models in Spark

Unit 8
Working with
LLMs in
Databricks
Enhance SQL operations using LLM’s
Databricks LLMs via SQL
- Databricks allows us to access LLM functionality right from SQL
- Accessible via calling the
ai_family of functions - New way of leveraging LLMs, batch scoring
https://docs.databricks.com/en/large-language-models/ai-functions-example.html
Call ai functions in dplyr


- Call the
aifunctions viadplyrverbs dbplyrpasses unrecognized functions ‘as-is’ in the query
orders |>
head() |>
select(o_comment) |>
mutate(
sentiment = ai_analyze_sentiment(o_comment)
)
#> # Source: SQL [6 x 2]
#> o_comment sentiment
#> <chr> <chr>
#> 1 ", pending theodolites … neutral
#> 2 "uriously special foxes … neutral
#> 3 "sleep. courts after the … neutral
#> 4 "ess foxes may sleep … neutral
#> 5 "ts wake blithely unusual … mixed
#> 6 "hins sleep. fluffily … neutralAvailable LLM functions
ai_analyze_sentiment- Text is positve, negative, neutral, etc.ai_classify- Give choices, LLM chooses which of the choice matchai_extract- Have LLM extract specific elements from the textai_fix_grammar- Makes grammar more betterai_mask- Replaces specified elements with ‘[Masked]’ai_similarity- Compares 2 texts, returns integer indicating similarityai_summarize- Summarizes text, you can also specify length of outputai_translate- Tell LLM to translate to specific languageai_forecast- Time series prediction, not text
Considerations in Databricks
- Need APIs pay-per-token supported region
- Functions not available on Databricks SQL Classic
- In preview, functions have performance restrictions
- Functions are tuned for English, but model can handle several languages
- Mixtral-8x7B Instruct is the underlying model that powers most of these AI functions
Exercise 8.1
Specify array
- Most
aifunctions require an argument ofARRAYtype - SQL’s
array()function can be called directly inside theaifunction
Exercise 8.2
Process complex output
- Some
aifunctions return aSTRUCT - It is a more complex variable type
- It can only be coerced to a string
as.character()does the trick
tbl_reviews |>
mutate(
product = as.character(
ai_extract(review, array("product"))
)
) |>
select(product, review)
#> # Source: SQL [4 x 2]
#> product review
#> <chr> <chr>
#> 1 {toaster} This is the best toaster I have ever bought
#> 2 {Toaster} Toaster arrived broken, waiting for replancement
#> 3 {washing machine} The washing machine is as advertised, cant wait to use it
#> 4 {television} Not sure how to feel about this tevelision, nice brightness…Exercise 8.3
Chat with LLM’s from R
Introducing chattr

chattr supports Databricks’ LLM

chattr supports Databricks’ LLM

Big thank you to Zack Davies at Databricks!! 🎉🎉🎉

https://github.com/mlverse/chattr/pull/99
chattr supports Databricks’ LLM

Automatically makes the options available if it detects your Databricks token.
> chattr_app()
── chattr - Available models
Select the number of the model you would like to use:
1: Databricks - databricks-dbrx-instruct (databricks-dbrx)
2: Databricks - databricks-meta-llama-3-70b-instruct (databricks-meta-llama3-70b)
3: Databricks - databricks-mixtral-8x7b-instruct (databricks-mixtral8x7b)
Selection: 1Exercise 8.4
Unit 9
Dashboards
Why dashboards?
🔑 Two fundamental points:
- App load time matters! The faster the better!
- Download raw data only if there is no other option
Important features
- Data driven dropdowns
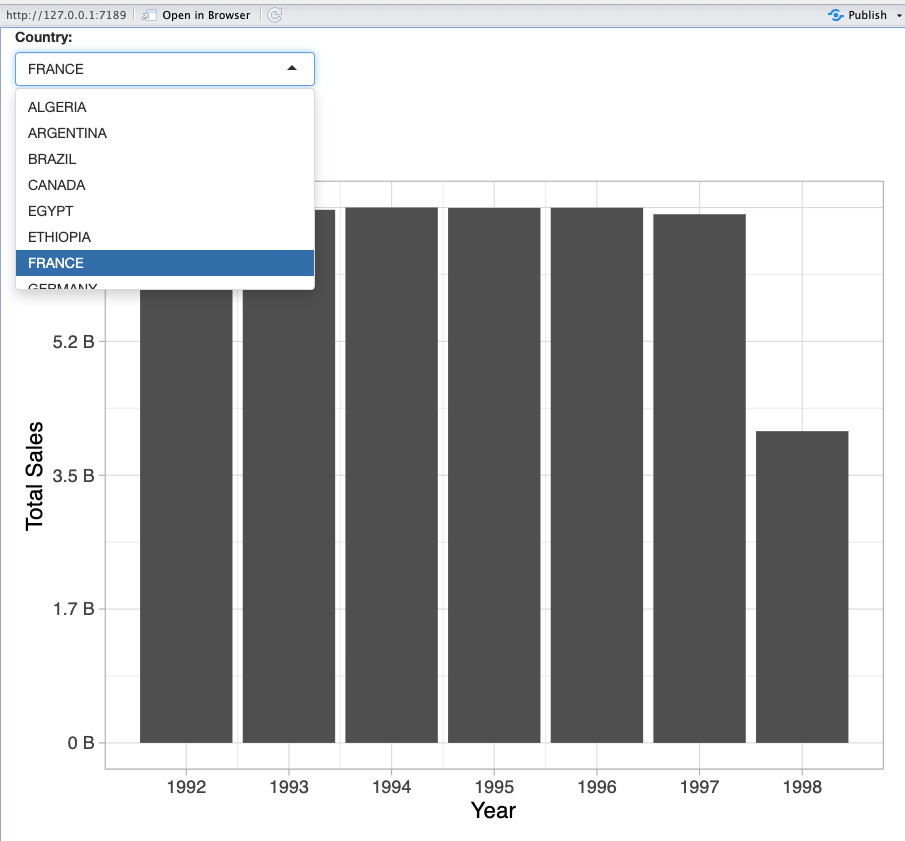
Important features
- Data driven dropdowns
- Interactive plots (hover over)
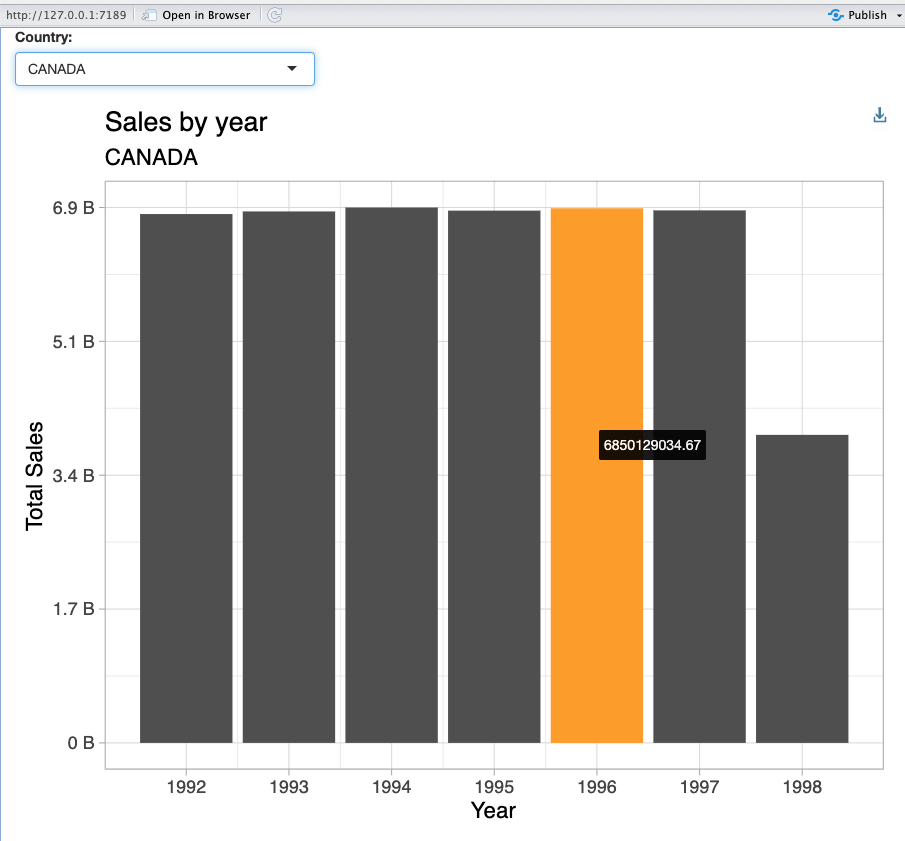
Important features
- Data driven dropdowns
- Interactive plots (hover over)
- Drill down when interacting with the plot
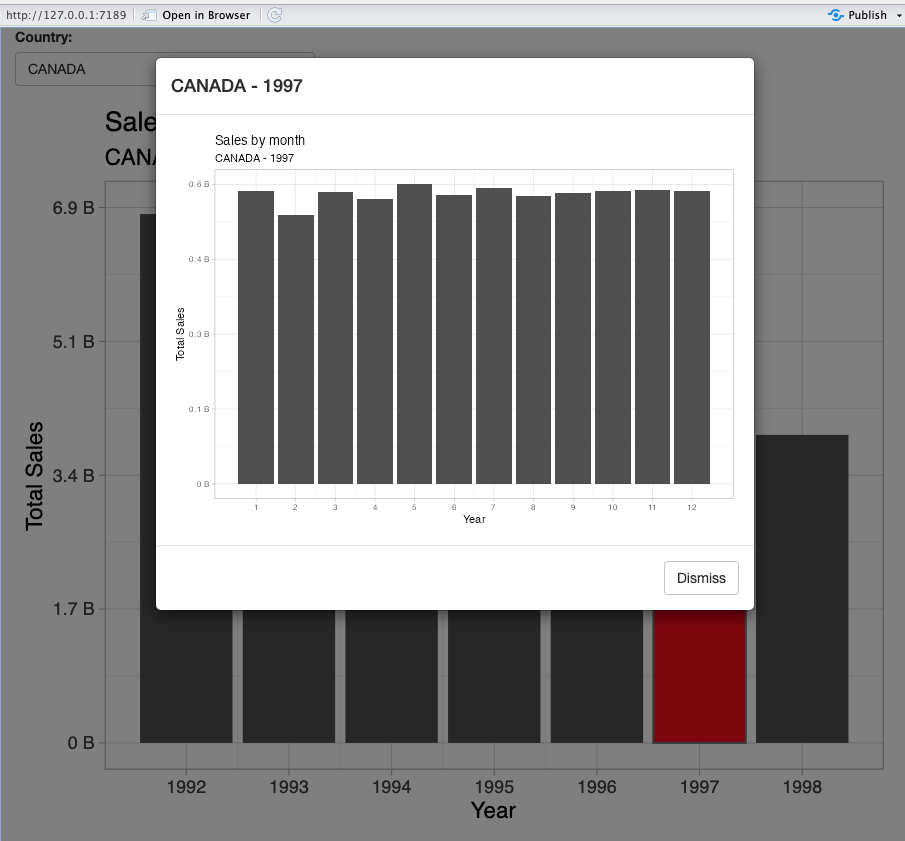
Exercise 9.1
Data driven dropdowns

When do we need them?
When options in the data change regularly
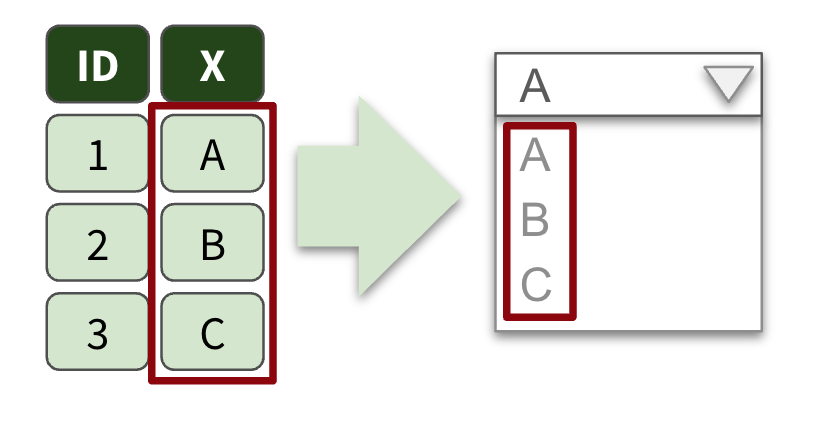
Exercise 9.2
Adding the plot
Move the code from the previous unit to the app
sales_by_year <- prep_orders |>
filter(n_name == country) |>
group_by(order_year) |>
summarise(
total_price = sum(o_totalprice, na.rm = TRUE)
) |>
collect()
breaks <- as.double(quantile(c(0, max(sales_by_year$total_price))))
breaks_labels <- paste(round(breaks / 1000000000, 1), "B")
sales_by_year |>
ggplot() +
geom_col(aes(order_year, total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = country) +
theme_light()Adding the plot
Prefix input$ to both instances of country
sales_by_year <- prep_orders |>
filter(n_name == country) |>
group_by(order_year) |>
summarise(total_price = sum(o_totalprice, na.rm = TRUE)) |>
collect()
sales_by_year |>
ggplot() +
geom_col(aes(order_year, total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = country) +
theme_light()Adding the plot
Prefix input$ to both instances of country
sales_by_year <- prep_orders |>
filter(n_name == input$country) |>
group_by(order_year) |>
summarise(total_price = sum(o_totalprice, na.rm = TRUE)) |>
collect()
sales_by_year |>
ggplot() +
geom_col(aes(order_year, total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = input$country) +
theme_light()Exercise 9.3
Interactive Plots

ggiraph

- Allows you to create dynamic
ggplotgraphs. - Adds tooltips, hover effects, and JavaScript actions
- Enables selecting elements of a plot inside a
shinyapp - Adds interactivity to
ggplotgeometries, legends and theme elements
https://davidgohel.github.io/ggiraph/
Interactive Plots
Replace with ggiraph functions
ui = fluidPage(
selectInput("country", "Country:", choices = countries, selected = "FRANCE"),
plotOutput()("sales_plot")),
server = function(input, output) {
output$sales_plot <- renderPlot()({
sales_by_year <- prep_orders |>
filter(n_name == !!input$country) |>
group_by(order_year) |>
summarise(total_price = sum(o_totalprice, na.rm = TRUE)) |>
collect()
sales_by_year |>
ggplot() +
geom_col(aes(order_year, total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +Interactive Plots
Replace with ggiraph functions
ui = fluidPage(
selectInput("country", "Country:", choices = countries, selected = "FRANCE"),
girafeOutput("sales_plot")),
server = function(input, output) {
output$sales_plot <- renderGirafe({
sales_by_year <- prep_orders |>
filter(n_name == !!input$country) |>
group_by(order_year) |>
summarise(total_price = sum(o_totalprice, na.rm = TRUE)) |>
collect()
sales_by_year |>
ggplot() +
geom_col_interactive(aes(order_year, total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +Interactive Plots
Add girafe() output step
sales_by_year |>
ggplot() +
geom_col_interactive(aes(order_year, total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = input$country) +
theme_light()Interactive Plots
Add girafe() output step
g <- sales_by_year |>
ggplot() +
geom_col_interactive(aes(order_year, total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = input$country) +
theme_light()
girafe(ggobj = g)Exercise 9.4
Plot drill-down

Plot drill down
Add a showModal() when a column is clicked
g <- sales_by_year |>
ggplot() +
geom_col_interactive(aes(order_year, total_price, data_id = order_year, tooltip = total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = input$country) +
theme_light()
girafe(ggobj = g)
})
},
options = list(height = 200)Plot drill down
Add a showModal() when a column is clicked
g <- sales_by_year |>
ggplot() +
geom_col_interactive(aes(order_year, total_price, data_id = order_year, tooltip = total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = input$country) +
theme_light()
girafe(ggobj = g)
})
observeEvent(input$sales_plot_selected, {
showModal(modalDialog())
})
},
options = list(height = 200)Plot drill down
Allow only 1 column to be selected at the time
g <- sales_by_year |>
ggplot() +
geom_col_interactive(aes(order_year, total_price, data_id = order_year, tooltip = total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = input$country) +
theme_light()
girafe(ggobj = g)
})
observeEvent(input$sales_plot_selected, {
showModal(modalDialog())
})
},
options = list(height = 200)Plot drill down
Allow only 1 column to be selected at the time
g <- sales_by_year |>
ggplot() +
geom_col_interactive(aes(order_year, total_price, data_id = order_year, tooltip = total_price)) +
scale_x_continuous(breaks = unique(sales_by_year$order_year)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(title = "Sales by year", subtitle = input$country) +
theme_light()
girafe(ggobj = g, options = list(opts_selection(type = "single")))
})
observeEvent(input$sales_plot_selected, {
showModal(modalDialog())
})
},
options = list(height = 200)Plot drill down
Add a dynamic title using the country and year selected
Plot drill down
Add a dynamic title using the country and year selected
Plot drill down
Add a new renderPLot() function, with code from Plot data by month
output$monthly_sales <- renderPlot({
sales_by_month <- prep_orders |>
filter(
n_name == !!input$country,
order_year == !!input$sales_plot_selected
) |>
group_by(order_month) |>
summarise(
total_price = sum(o_totalprice, na.rm = TRUE)
) |>
collect()
breaks <- as.double(quantile(c(0, max(sales_by_month$total_price))))
breaks_labels <- paste(round(breaks / 1000000000, 1), "B")
sales_by_month |>
ggplot() +
geom_col(aes(order_month, total_price)) +
scale_x_continuous(breaks = unique(sales_by_month$order_month)) +
scale_y_continuous(breaks = breaks, labels = breaks_labels) +
xlab("Year") +
ylab("Total Sales") +
labs(
title = "Sales by month",
subtitle = paste0(input$country, " - ", input$sales_plot_selected)
) +
theme_light()
})Plot drill down
Add plotOutput("monthly_sales") to the modelDialog() call
Plot drill down
Add plotOutput("monthly_sales") to the modelDialog() call
Additional resources
Official Site
spark.posit.co
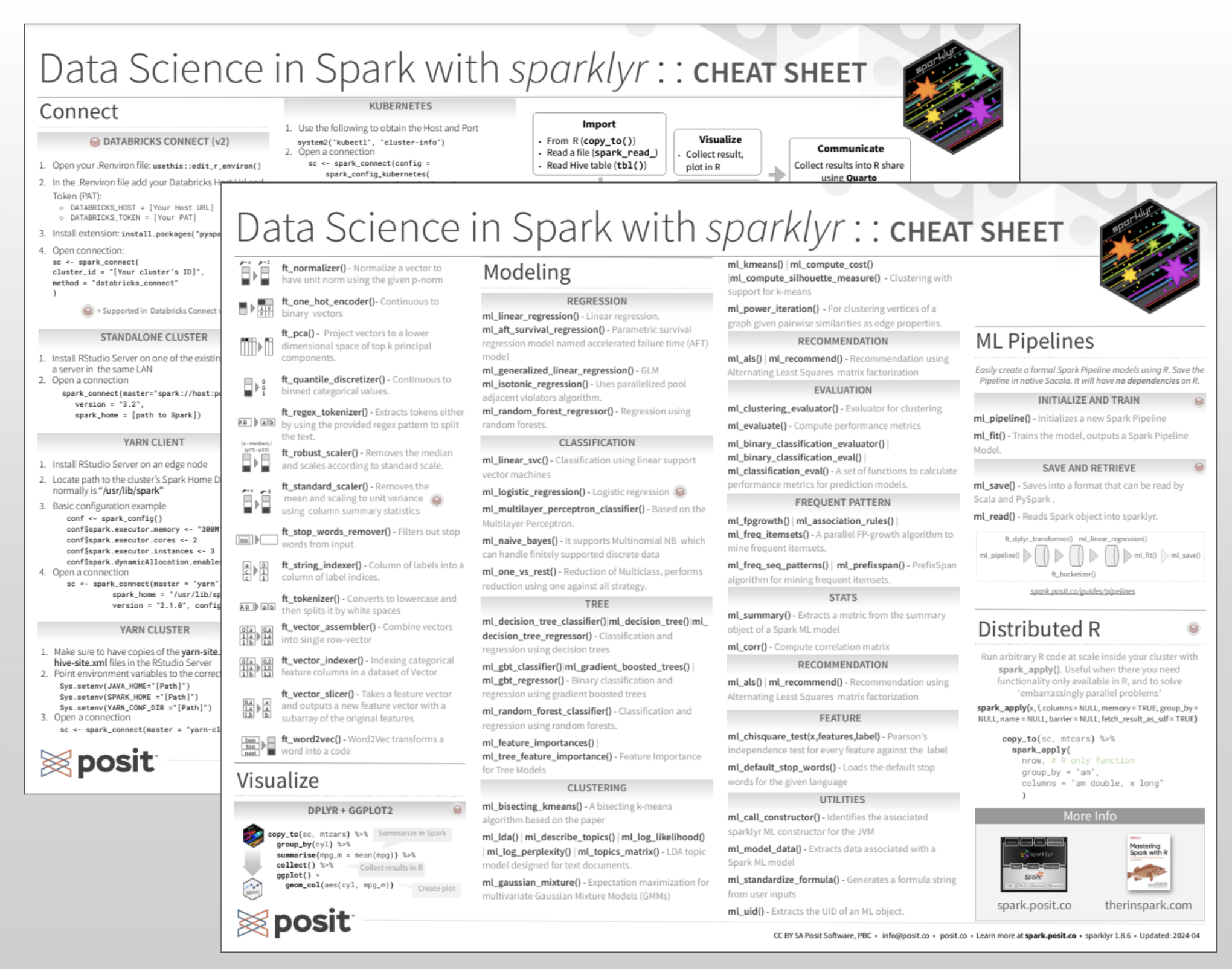
Cheatsheet
spark.posit.co/images/homepage/sparklyr.pdf
Ask questions
forum.posit.co
Let us know what you thought
Please take our survey:
pos.it/conf-workshop-survey
