<class 'pandas.core.frame.DataFrame'>
Index: 303 entries, 1 to 303
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Age 303 non-null int64
1 Sex 303 non-null int64
2 ChestPain 303 non-null object
3 RestBP 303 non-null int64
4 Chol 303 non-null int64
5 Fbs 303 non-null int64
6 RestECG 303 non-null int64
7 MaxHR 303 non-null int64
8 ExAng 303 non-null int64
9 Oldpeak 303 non-null float64
10 Slope 303 non-null int64
11 Ca 299 non-null float64
12 Thal 301 non-null object
13 AHD 303 non-null object
dtypes: float64(2), int64(9), object(3)
memory usage: 35.5+ KBTree-based and ensemble models
Tree-based methods
Algorithms that stratifying or segmenting the predictor space into a number of simple regions.
We call these algorithms decision-tree methods because the decisions used to segment the predictor space can be summarized in a tree.
Decision trees on their own, are very explainable and intuitive, but not very powerful at predicting.
However, there are extensions of decision trees, such as random forest and boosted trees, which are very powerful at predicting. We will demonstrate two of these in this session.
Decision trees
- Decision Trees by Jared Wilber & Lucía Santamaría
Classification Decision trees
Use recursive binary splitting to grow a classification tree (splitting of the predictor space into \(J\) distinct, non-overlapping regions).
For every observation that falls into the region \(R_j\) , we make the same prediction, which is the majority vote for the training observations in \(R_j\).
Where to split the predictor space is done in a top-down and greedy manner, and in practice for classification, the best split at any point in the algorithm is one that minimizes the Gini index (a measure of node purity).
Decision trees are useful because they are very interpretable.
A limitation of decision trees is that theyn tend to overfit, so in practice we use cross-validation to tune a hyperparameter, \(\alpha\), to find the optimal, pruned tree.
Example: the heart data set
Let’s consider a situation where we’d like to be able to predict the presence of heart disease (
AHD) in patients, based off 13 measured characteristics.The heart data set contains a binary outcome for heart disease for patients who presented with chest pain.
Example: the heart data set
An angiographic test was performed and a label for AHD of Yes was labelled to indicate the presence of heart disease, otherwise the label was No.
| Age | Sex | ChestPain | RestBP | Chol | Fbs | RestECG | MaxHR | ExAng | Oldpeak | Slope | Ca | Thal | AHD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 63 | 1 | typical | 145 | 233 | 1 | 2 | 150 | 0 | 2.3 | 3 | 0.0 | fixed | No |
| 2 | 67 | 1 | asymptomatic | 160 | 286 | 0 | 2 | 108 | 1 | 1.5 | 2 | 3.0 | normal | Yes |
| 3 | 67 | 1 | asymptomatic | 120 | 229 | 0 | 2 | 129 | 1 | 2.6 | 2 | 2.0 | reversable | Yes |
| 4 | 37 | 1 | nonanginal | 130 | 250 | 0 | 0 | 187 | 0 | 3.5 | 3 | 0.0 | normal | No |
| 5 | 41 | 0 | nontypical | 130 | 204 | 0 | 2 | 172 | 0 | 1.4 | 1 | 0.0 | normal | No |
Do we have a class imbalance?
It’s always important to check this, as it may impact your splitting and/or modeling decisions.
AHD
No 0.541254
Yes 0.458746
Name: proportion, dtype: float64This looks pretty good! We can move forward this time without doing much more about this.
Data splitting
Let’s split the data into training and test sets:
import numpy as np
from sklearn.model_selection import train_test_split
np.random.seed(2024)
heart_train, heart_test = train_test_split(
heart, train_size=0.8, stratify=heart["AHD"]
)
X_train = heart_train.drop(columns=['AHD'])
y_train = heart_train['AHD']
X_test = heart_test.drop(columns=['AHD'])
y_test = heart_test['AHD']Categorical variables
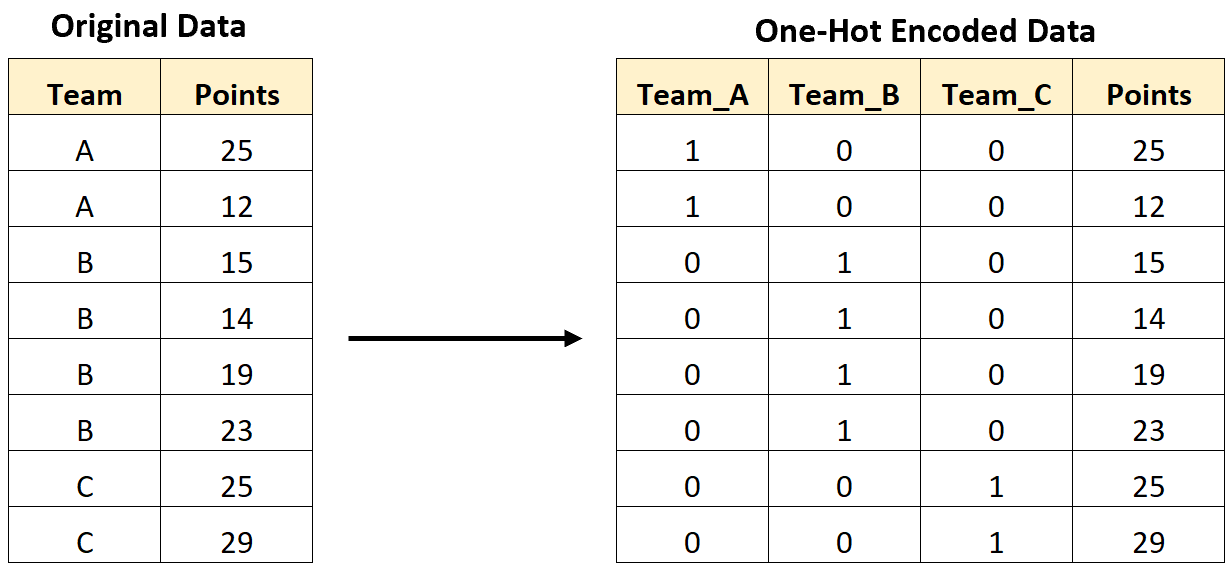
This is our first case of seeing categorical predictor variables, can we treat them the same as numerical ones? No!
In
scikit-learnwe must perform one-hot encoding
Look at the data again
Which columns do we need to standardize?
Which do we need to one-hot encode?
| Age | Sex | ChestPain | RestBP | Chol | Fbs | RestECG | MaxHR | ExAng | Oldpeak | Slope | Ca | Thal | AHD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 63 | 1 | typical | 145 | 233 | 1 | 2 | 150 | 0 | 2.3 | 3 | 0.0 | fixed | No |
| 2 | 67 | 1 | asymptomatic | 160 | 286 | 0 | 2 | 108 | 1 | 1.5 | 2 | 3.0 | normal | Yes |
| 3 | 67 | 1 | asymptomatic | 120 | 229 | 0 | 2 | 129 | 1 | 2.6 | 2 | 2.0 | reversable | Yes |
| 4 | 37 | 1 | nonanginal | 130 | 250 | 0 | 0 | 187 | 0 | 3.5 | 3 | 0.0 | normal | No |
| 5 | 41 | 0 | nontypical | 130 | 204 | 0 | 2 | 172 | 0 | 1.4 | 1 | 0.0 | normal | No |
One hot encoding & pre-processing
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.compose import make_column_transformer, make_column_selector
numeric_feats = ['Age', 'RestBP', 'Chol', 'RestECG', 'MaxHR', 'Oldpeak','Slope', 'Ca']
passthrough_feats = ['Sex', 'Fbs', 'ExAng']
categorical_feats = ['ChestPain', 'Thal']
heart_preprocessor = make_column_transformer(
(StandardScaler(), numeric_feats),
("passthrough", passthrough_feats),
(OneHotEncoder(handle_unknown = "ignore"), categorical_feats),
)
handle_unknown = "ignore"handles the case where categories exist in the test data, which were missing in the training set. Specifically, it sets the value for those to 0 for all cases of the category.
Fitting a dummy classifier
from sklearn.dummy import DummyClassifier
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import cross_validate
dummy = DummyClassifier()
dummy_pipeline = make_pipeline(heart_preprocessor, dummy)
cv_10_dummy = pd.DataFrame(
cross_validate(
estimator=dummy_pipeline,
cv=10,
X=X_train,
y=y_train
)
)
cv_10_dummy_metrics = cv_10_dummy.agg(["mean", "sem"])
results = pd.DataFrame({'mean' : [cv_10_dummy_metrics.test_score.iloc[0]],
'sem' : [cv_10_dummy_metrics.test_score.iloc[1]]},
index = ['Dummy classifier']
)
results| mean | sem | |
|---|---|---|
| Dummy classifier | 0.541333 | 0.00299 |
Fitting a decision tree
from sklearn.tree import DecisionTreeClassifier
decision_tree = DecisionTreeClassifier(random_state=2026)
dt_pipeline = make_pipeline(heart_preprocessor, decision_tree)
cv_10_dt = pd.DataFrame(
cross_validate(
estimator=dt_pipeline,
cv=10,
X=X_train,
y=y_train
)
)
cv_10_dt_metrics = cv_10_dt.agg(["mean", "sem"])
results_dt = pd.DataFrame({'mean' : [cv_10_dt_metrics.test_score.iloc[0]],
'sem' : [cv_10_dt_metrics.test_score.iloc[1]]},
index = ['Decision tree']
)
results = pd.concat([results, results_dt])
results| mean | sem | |
|---|---|---|
| Dummy classifier | 0.541333 | 0.00299 |
| Decision tree | 0.769167 | 0.02632 |
Can we do better?
We could tune some decision tree parameters (e.g., alpha, maximum tree depth, etc)…
We could also try a different tree-based method!
The Random Forest Algorithm by Jenny Yeon & Jared Wilber
The Random Forest Algorithm
Build a number of decision trees on bootstrapped training samples.
When building the trees from the bootstrapped samples, at each stage of splitting, the best splitting is computed using a randomly selected subset of the features.
Take the majority votes across all the trees for the final prediction.
Random forest in scikit-learn & missing values
Does not accept missing values, we need to deal with these somehow…
We can either drop the observations with missing values, or we can somehow impute them.
For the purposes of this demo we will drop them, but if you are interested in imputation, see the imputation tutorial in
scikit-learn
Random forest in scikit-learn
from sklearn.ensemble import RandomForestClassifier
random_forest = RandomForestClassifier(random_state=2026)
rf_pipeline = make_pipeline(heart_preprocessor, random_forest)
cv_10_rf = pd.DataFrame(
cross_validate(
estimator=rf_pipeline,
cv=10,
X=X_train_drop_na,
y=y_train_drop_na
)
)
cv_10_rf_metrics = cv_10_rf.agg(["mean", "sem"])
results_rf = pd.DataFrame({'mean' : [cv_10_rf_metrics.test_score.iloc[0]],
'sem' : [cv_10_rf_metrics.test_score.iloc[1]]},
index = ['Random forest']
)
results = pd.concat([results, results_rf])
results| mean | sem | |
|---|---|---|
| Dummy classifier | 0.541333 | 0.002990 |
| Decision tree | 0.769167 | 0.026320 |
| Random forest | 0.818297 | 0.017362 |
Can we do better?
Random forest can be tuned a several important parameters, including:
n_estimators: number of decision trees (higher = more complexity)max_depth: max depth of each decision tree (higher = more complexity)max_features: the number of features you get to look at each split (higher = more complexity)
We can use
GridSearchCVto search for the optimal parameters for these, as we did for \(K\) in \(K\)-nearest neighbors.
Tuning random forest in scikit-learn
from sklearn.model_selection import GridSearchCV
rf_param_grid = {'randomforestclassifier__n_estimators': [200],
'randomforestclassifier__max_depth': [1, 3, 5, 7, 9],
'randomforestclassifier__max_features': [1, 2, 3, 4, 5, 6, 7]}
rf_tune_grid = GridSearchCV(
estimator=rf_pipeline,
param_grid=rf_param_grid,
cv=10,
n_jobs=-1 # tells computer to use all available CPUs
)
rf_tune_grid.fit(
X_train_drop_na,
y_train_drop_na
)
cv_10_rf_tuned_metrics = pd.DataFrame(rf_tune_grid.cv_results_)
results_rf_tuned = pd.DataFrame({'mean' : rf_tune_grid.best_score_,
'sem' : pd.DataFrame(rf_tune_grid.cv_results_)['std_test_score'][6] / 10**(1/2)},
index = ['Random forest tuned']
)
results = pd.concat([results, results_rf_tuned])Random Forest results
How did the Random Forest compare against the other models we tried?
Boosting
No randomization.
The key idea is combining many simple models called weak learners, to create a strong learner.
They combine multiple shallow (depth 1 to 5) decision trees.
They build trees in a serial manner, where each tree tries to correct the mistakes of the previous one.
Tuning GradientBoostingClassifier with scikit-learn
GradientBoostingClassifiercan be tuned a several important parameters, including:n_estimators: number of decision trees (higher = more complexity)max_depth: max depth of each decision tree (higher = more complexity)learning_rate: the shrinkage parameter which controls the rate at which boosting learns. Values between 0.01 or 0.001 are typical.
We can use
GridSearchCVto search for the optimal parameters for these, as we did for the parameters in Random Forest.
Tuning GradientBoostingClassifier with scikit-learn
from sklearn.ensemble import GradientBoostingClassifier
gradient_boosted_classifier = GradientBoostingClassifier(random_state=2026)
gb_pipeline = make_pipeline(heart_preprocessor, gradient_boosted_classifier)
gb_param_grid = {'gradientboostingclassifier__n_estimators': [200],
'gradientboostingclassifier__max_depth': [1, 3, 5, 7, 9],
'gradientboostingclassifier__learning_rate': [0.001, 0.005, 0.01]}
gb_tune_grid = GridSearchCV(
estimator=gb_pipeline,
param_grid=gb_param_grid,
cv=10,
n_jobs=-1 # tells computer to use all available CPUs
)
gb_tune_grid.fit(
X_train_drop_na,
y_train_drop_na
)
cv_10_gb_tuned_metrics = pd.DataFrame(gb_tune_grid.cv_results_)
results_gb_tuned = pd.DataFrame({'mean' : gb_tune_grid.best_score_,
'sem' : pd.DataFrame(gb_tune_grid.cv_results_)['std_test_score'][6] / 10**(1/2)},
index = ['Gradient boosted classifier tuned']
)
results = pd.concat([results, results_gb_tuned])GradientBoostingClassifier results
How did the GradientBoostingClassifier compare against the other models we tried?
How do we choose the final model?
Remember, what is your question or application?
A good rule when models are not very different, what is the simplest model that does well?
Look at other metrics that are important to you (not just the metric you used for tuning your model), remember precision & recall, for example.
Remember - no peaking at the test set until you choose! And then, you should only look at the test set for one model!
Precision and recall on the tuned random forest model
from sklearn.metrics import make_scorer, precision_score, recall_score
scoring = {
'accuracy': 'accuracy',
'precision': make_scorer(precision_score, pos_label='Yes'),
'recall': make_scorer(recall_score, pos_label='Yes')
}
rf_tune_grid = GridSearchCV(
estimator=rf_pipeline,
param_grid=rf_param_grid,
cv=10,
n_jobs=-1,
scoring=scoring,
refit='accuracy'
)
rf_tune_grid.fit(X_train_drop_na, y_train_drop_na)GridSearchCV(cv=10,
estimator=Pipeline(steps=[('columntransformer',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['Age',
'RestBP',
'Chol',
'RestECG',
'MaxHR',
'Oldpeak',
'Slope',
'Ca']),
('passthrough',
'passthrough',
['Sex',
'Fbs',
'ExAng']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['ChestPain',
'Thal'])])),
('randomforestclas...
param_grid={'randomforestclassifier__max_depth': [1, 3, 5, 7, 9],
'randomforestclassifier__max_features': [1, 2, 3, 4, 5,
6, 7],
'randomforestclassifier__n_estimators': [200]},
refit='accuracy',
scoring={'accuracy': 'accuracy',
'precision': make_scorer(precision_score, response_method='predict', pos_label=Yes),
'recall': make_scorer(recall_score, response_method='predict', pos_label=Yes)})In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(cv=10,
estimator=Pipeline(steps=[('columntransformer',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['Age',
'RestBP',
'Chol',
'RestECG',
'MaxHR',
'Oldpeak',
'Slope',
'Ca']),
('passthrough',
'passthrough',
['Sex',
'Fbs',
'ExAng']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['ChestPain',
'Thal'])])),
('randomforestclas...
param_grid={'randomforestclassifier__max_depth': [1, 3, 5, 7, 9],
'randomforestclassifier__max_features': [1, 2, 3, 4, 5,
6, 7],
'randomforestclassifier__n_estimators': [200]},
refit='accuracy',
scoring={'accuracy': 'accuracy',
'precision': make_scorer(precision_score, response_method='predict', pos_label=Yes),
'recall': make_scorer(recall_score, response_method='predict', pos_label=Yes)})Pipeline(steps=[('columntransformer',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['Age', 'RestBP', 'Chol',
'RestECG', 'MaxHR',
'Oldpeak', 'Slope', 'Ca']),
('passthrough', 'passthrough',
['Sex', 'Fbs', 'ExAng']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['ChestPain', 'Thal'])])),
('randomforestclassifier',
RandomForestClassifier(max_depth=1, max_features=2,
n_estimators=200, random_state=2026))])ColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['Age', 'RestBP', 'Chol', 'RestECG', 'MaxHR',
'Oldpeak', 'Slope', 'Ca']),
('passthrough', 'passthrough',
['Sex', 'Fbs', 'ExAng']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['ChestPain', 'Thal'])])['Age', 'RestBP', 'Chol', 'RestECG', 'MaxHR', 'Oldpeak', 'Slope', 'Ca']
StandardScaler()
['Sex', 'Fbs', 'ExAng']
passthrough
['ChestPain', 'Thal']
OneHotEncoder(handle_unknown='ignore')
RandomForestClassifier(max_depth=1, max_features=2, n_estimators=200,
random_state=2026)Precision and recall cont’d
What do we think? Is this model ready for production in a diagnostic setting?
How could we improve it further?
cv_results = pd.DataFrame(rf_tune_grid.cv_results_)
mean_precision = cv_results['mean_test_precision'].iloc[rf_tune_grid.best_index_]
sem_precision = cv_results['std_test_precision'].iloc[rf_tune_grid.best_index_] / np.sqrt(10)
mean_recall = cv_results['mean_test_recall'].iloc[rf_tune_grid.best_index_]
sem_recall = cv_results['std_test_recall'].iloc[rf_tune_grid.best_index_] / np.sqrt(10)
results_rf_tuned = pd.DataFrame({
'mean': [rf_tune_grid.best_score_, mean_precision, mean_recall],
'sem': [cv_results['std_test_accuracy'].iloc[rf_tune_grid.best_index_] / np.sqrt(10), sem_precision, sem_recall],
}, index=['accuracy', 'precision', 'recall'])
results_rf_tuned| mean | sem | |
|---|---|---|
| accuracy | 0.860688 | 0.018576 |
| precision | 0.920505 | 0.022982 |
| recall | 0.770909 | 0.038928 |
Feature importances
Key points:
Decision trees are very interpretable (decision rules!), however in ensemble models (e.g., Random Forest and Boosting) there are many trees - individual decision rules are not as meaningful…
Instead, we can calculate feature importances as the total decrease in impurity for all splits involving that feature, weighted by the number of samples involved in those splits, normalized and averaged over all the trees.
These are calculated on the training set, as that is the set the model is trained on.
Notes of caution!
Feature importances can be unreliable with both highly cardinal, and multicollinear features.
Unlike the linear model coefficients, feature importances do not have a sign! They tell us about importance, but not an “up or down”.
Increasing a feature may cause the prediction to first go up, and then go down.
Alternatives to feature importance to understanding models exist (e.g., SHAP (SHapley Additive exPlanations))
Feature importances in scikit-learn
# Access the best pipeline
best_pipeline = rf_tune_grid.best_estimator_
# Extract the trained RandomForestClassifier from the pipeline
best_rf = best_pipeline.named_steps['randomforestclassifier']
# Extract feature names after preprocessing
# Get the names of features from each transformer in the pipeline
numeric_features = numeric_feats
categorical_feature_names = best_pipeline.named_steps['columntransformer'].transformers_[2][1].get_feature_names_out(categorical_feats)
passthrough_features = passthrough_feats
# Combine all feature names into a single list
feature_names = np.concatenate([numeric_features, passthrough_features, categorical_feature_names])
# Calculate feature importances
feature_importances = best_rf.feature_importances_
# Create a DataFrame to display feature importances
importances_df = pd.DataFrame({
'Feature': feature_names,
'Importance': feature_importances
})
# Sort by importance (descending order)
importances_df = importances_df.sort_values(by='Importance', ascending=False)Visualizing the results
Evaluating on the test set
Predict on the test set:
Evaluating on the test set
Examine accuracy, precision and recall:
Other boosting models:
- Not part of
sklearnbut has similar interface. - Supports missing values
- GPU training, networked parallel training
- Supports sparse data
- Typically better scores than random forests
Keep learning!
Local installation
Using Docker: Data Science: A First Introduction (Python Edition) Installation Instructions
Using conda: UBC MDS Installation Instructions
Additional resources
- The UBC DSCI 573 (Feature and Model Selection notes) chapter of Data Science: A First Introduction (Python Edition) by Varada Kolhatkar and Joel Ostblom. These notes cover classification and regression metrics, advanced variable selection and more on ensembles.
- The
scikit-learnwebsite is an excellent reference for more details on, and advanced usage of, the functions and packages in the past two chapters. Aside from that, it also offers many useful tutorials to get you started. - An Introduction to Statistical Learning {cite:p}
james2013introductionprovides a great next stop in the process of learning about classification. Chapter 4 discusses additional basic techniques for classification that we do not cover, such as logistic regression, linear discriminant analysis, and naive Bayes.
References
Gareth James, Daniela Witten, Trevor Hastie, Robert Tibshirani and Jonathan Taylor. An Introduction to Statistical Learning with Applications in Python. Springer, 1st edition, 2023. URL: https://www.statlearning.com/.
Kolhatkar, V., and Ostblom, J. (2024). UBC DSCI 573: Feature and Model Selection course notes. URL: https://ubc-mds.github.io/DSCI_573_feat-model-select
Pedregosa, F. et al., 2011. Scikit-learn: Machine learning in Python. Journal of machine learning research, 12(Oct), pp.2825–2830.