| Chemotherapy Treatment | Age | Marker Level (ng/mL) | T Stage | Grade | Tumor Response | Patient Died | Months to Death/Censor |
|---|---|---|---|---|---|---|---|
| Drug A | 23 | 0.160 | T1 | II | 0 | 0 | 24.00 |
| Drug B | 9 | 1.107 | T2 | I | 1 | 0 | 24.00 |
| Drug A | 31 | 0.277 | T1 | II | 0 | 0 | 24.00 |
| Drug A | NA | 2.067 | T3 | III | 1 | 1 | 17.64 |
| Drug A | 51 | 2.767 | T4 | III | 1 | 1 | 16.43 |
| Drug B | 39 | 0.613 | T4 | I | 0 | 1 | 15.64 |
Pharmaverse Phlavors: gtsummary + ARD
Introduction
Acknowledgements
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (CC BY-SA4.0).
Daniel D. Sjoberg
Questions
Please ask questions at any time!

How it started
Began to address reproducible issues while working in academia
Goal was to build a package to summarize study results with code that was both simple and customizable
First release in May 2019
How it’s going
The stats
- 1,000,000+ installations from CRAN
- 1000+ GitHub stars
- 300+ contributors
- ~50 code contributors
Won the 2021 American Statistical Association (ASA) Innovation in Programming Award
Won the 2024 Posit Pharma Table Contest



{gtsummary} overview
- Create tabular summaries with sensible defaults but highly customizable
- Types of summaries:
- Demographic- or “Table 1”-types
- Cross-tabulation
- Regression models
- Survival data
- Survey data
- Custom tables
- Report statistics from {gtsummary} tables inline in R Markdown
- Stack and/or merge any table type
- Use themes to standardize across tables
- Choose from different print engines
{gtsummary} overview
For our workshop, we will focus on the following summary types as well as themes and print engines.
tbl_summary()tbl_cross()tbl_continuous()tbl_wide_summary()
Example Dataset
The
trialdata set is included with {gtsummary}Simulated data set of baseline characteristics for 200 patients who receive Drug A or Drug B
Variables were assigned labels using the
labelledpackage
Example Dataset
tbl_summary()
Basic tbl_summary()
Four types of summaries:
continuous,continuous2,categorical, anddichotomousStatistics are
median (IQR)for continuous,n (%)for categorical/dichotomousVariables coded
0/1,TRUE/FALSE,Yes/Notreated as dichotomousLists
NAvalues under “Unknown”Label attributes are printed automatically
Customize tbl_summary() output
| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|
| Age | 46 (37, 60) | 48 (39, 56) |
| Unknown | 7 | 4 |
| Grade | ||
| I | 35 (36%) | 33 (32%) |
| II | 32 (33%) | 36 (35%) |
| III | 31 (32%) | 33 (32%) |
| Tumor Response | 28 (29%) | 33 (34%) |
| Unknown | 3 | 4 |
| 1 Median (Q1, Q3); n (%) | ||
by: specify a column variable for cross-tabulation
Customize tbl_summary() output
| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|
| Age | ||
| Median (Q1, Q3) | 46 (37, 60) | 48 (39, 56) |
| Unknown | 7 | 4 |
| Grade | ||
| I | 35 (36%) | 33 (32%) |
| II | 32 (33%) | 36 (35%) |
| III | 31 (32%) | 33 (32%) |
| Tumor Response | 28 (29%) | 33 (34%) |
| Unknown | 3 | 4 |
| 1 n (%) | ||
by: specify a column variable for cross-tabulationtype: specify the summary type
Customize tbl_summary() output
| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|
| Age | ||
| Mean (SD) | 47 (15) | 47 (14) |
| Min, Max | 6, 78 | 9, 83 |
| Unknown | 7 | 4 |
| Grade | ||
| I | 35 (36%) | 33 (32%) |
| II | 32 (33%) | 36 (35%) |
| III | 31 (32%) | 33 (32%) |
| Tumor Response | 28 / 95 (29%) | 33 / 98 (34%) |
| Unknown | 3 | 4 |
| 1 n (%); n / N (%) | ||
by: specify a column variable for cross-tabulationtype: specify the summary typestatistic: customize the reported statistics
Customize tbl_summary() output
| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|
| Age | ||
| Mean (SD) | 47 (15) | 47 (14) |
| Min, Max | 6, 78 | 9, 83 |
| Unknown | 7 | 4 |
| Pathologic tumor grade | ||
| I | 35 (36%) | 33 (32%) |
| II | 32 (33%) | 36 (35%) |
| III | 31 (32%) | 33 (32%) |
| Tumor Response | 28 / 95 (29%) | 33 / 98 (34%) |
| Unknown | 3 | 4 |
| 1 n (%); n / N (%) | ||
by: specify a column variable for cross-tabulationtype: specify the summary typestatistic: customize the reported statistics
label: change or customize variable labels
Customize tbl_summary() output
| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|
| Age | ||
| Mean (SD) | 47 (14.7) | 47 (14.0) |
| Min, Max | 6, 78 | 9, 83 |
| Unknown | 7 | 4 |
| Pathologic tumor grade | ||
| I | 35 (36%) | 33 (32%) |
| II | 32 (33%) | 36 (35%) |
| III | 31 (32%) | 33 (32%) |
| Tumor Response | 28 / 95 (29%) | 33 / 98 (34%) |
| Unknown | 3 | 4 |
| 1 n (%); n / N (%) | ||
by: specify a column variable for cross-tabulationtype: specify the summary typestatistic: customize the reported statistics
label: change or customize variable labelsdigits: specify the number of decimal places for rounding
{gtsummary} + formulas
This syntax is also used in {cards}, {cardx}, and {gt}.
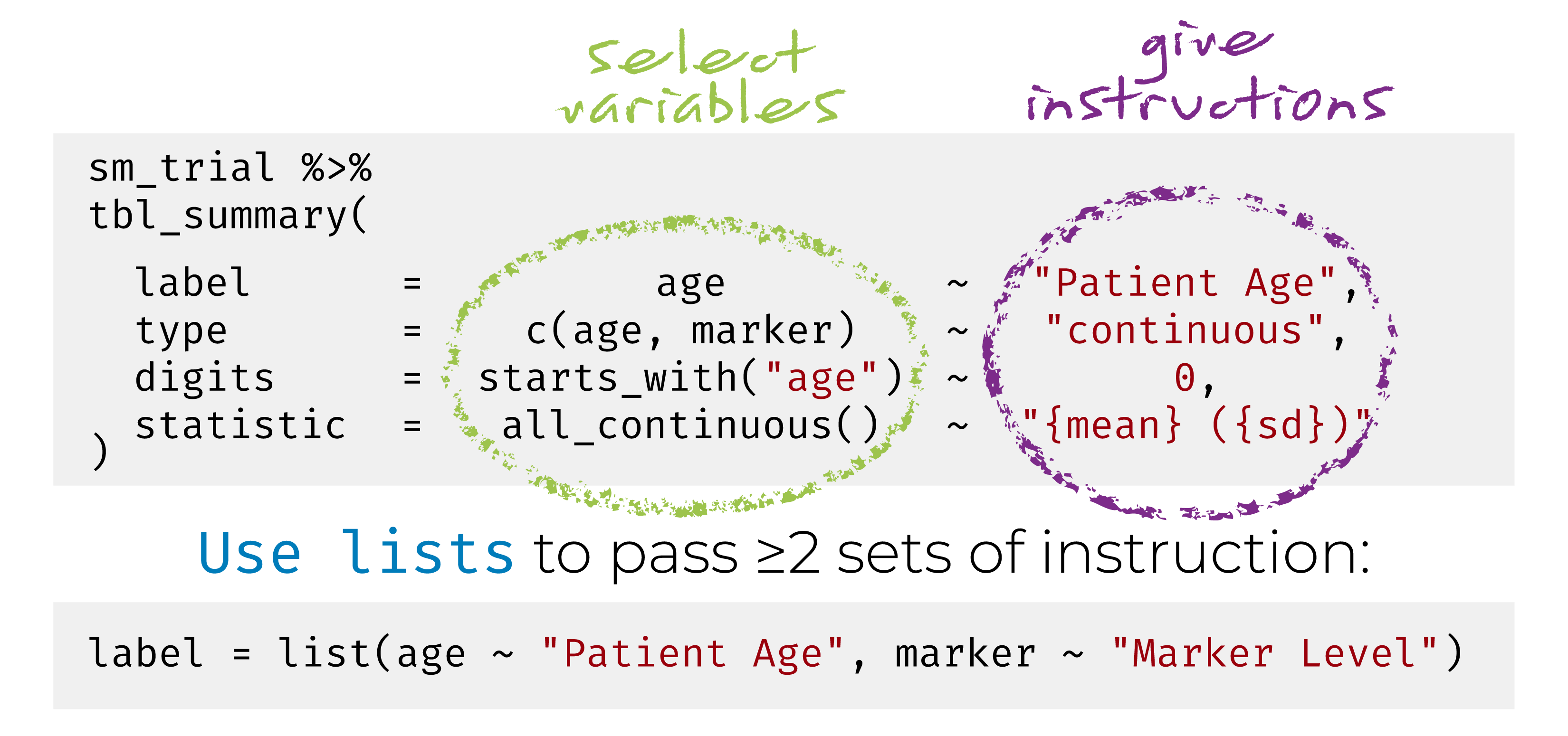
Named list are OK too! label = list(age = "Patient Age")
{gtsummary} selectors
Use the following helpers to select groups of variables:
all_continuous(),all_categorical()Use
all_stat_cols()to select the summary statistic columns
Add-on functions in {gtsummary}
tbl_summary() objects can also be updated using related functions.
add_*()add additional column of statistics or information, e.g. p-values, q-values, overall statistics, treatment differences, N obs., and moremodify_*()modify table headers, spanning headers, footnotes, and morebold_*()/italicize_*()style labels, variable levels, significant p-values
Update tbl_summary() with add_*()
| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
p-value2 | q-value3 |
|---|---|---|---|---|
| Age | 46 (37, 60) | 48 (39, 56) | 0.7 | 0.9 |
| Unknown | 7 | 4 | ||
| Grade | 0.9 | 0.9 | ||
| I | 35 (36%) | 33 (32%) | ||
| II | 32 (33%) | 36 (35%) | ||
| III | 31 (32%) | 33 (32%) | ||
| Tumor Response | 28 (29%) | 33 (34%) | 0.5 | 0.9 |
| Unknown | 3 | 4 | ||
| 1 Median (Q1, Q3); n (%) | ||||
| 2 Wilcoxon rank sum test; Pearson’s Chi-squared test | ||||
| 3 False discovery rate correction for multiple testing | ||||
add_p(): adds a column of p-valuesadd_q(): adds a column of p-values adjusted for multiple comparisons through a call top.adjust()
Update tbl_summary() with add_*()
| Characteristic | Overall N = 2001 |
Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|---|
| Age | 47 (38, 57) | 46 (37, 60) | 48 (39, 56) |
| Grade | |||
| I | 68 (34%) | 35 (36%) | 33 (32%) |
| II | 68 (34%) | 32 (33%) | 36 (35%) |
| III | 64 (32%) | 31 (32%) | 33 (32%) |
| Tumor Response | 61 (32%) | 28 (29%) | 33 (34%) |
| 1 Median (Q1, Q3); n (%) | |||
add_overall(): adds a column of overall statistics
Update tbl_summary() with add_*()
| Characteristic | N | Overall N = 2001 |
Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|---|---|
| Age | 189 | 47 (38, 57) | 46 (37, 60) | 48 (39, 56) |
| Grade | 200 | |||
| I | 68 (34%) | 35 (36%) | 33 (32%) | |
| II | 68 (34%) | 32 (33%) | 36 (35%) | |
| III | 64 (32%) | 31 (32%) | 33 (32%) | |
| Tumor Response | 193 | 61 (32%) | 28 (29%) | 33 (34%) |
| 1 Median (Q1, Q3); n (%) | ||||
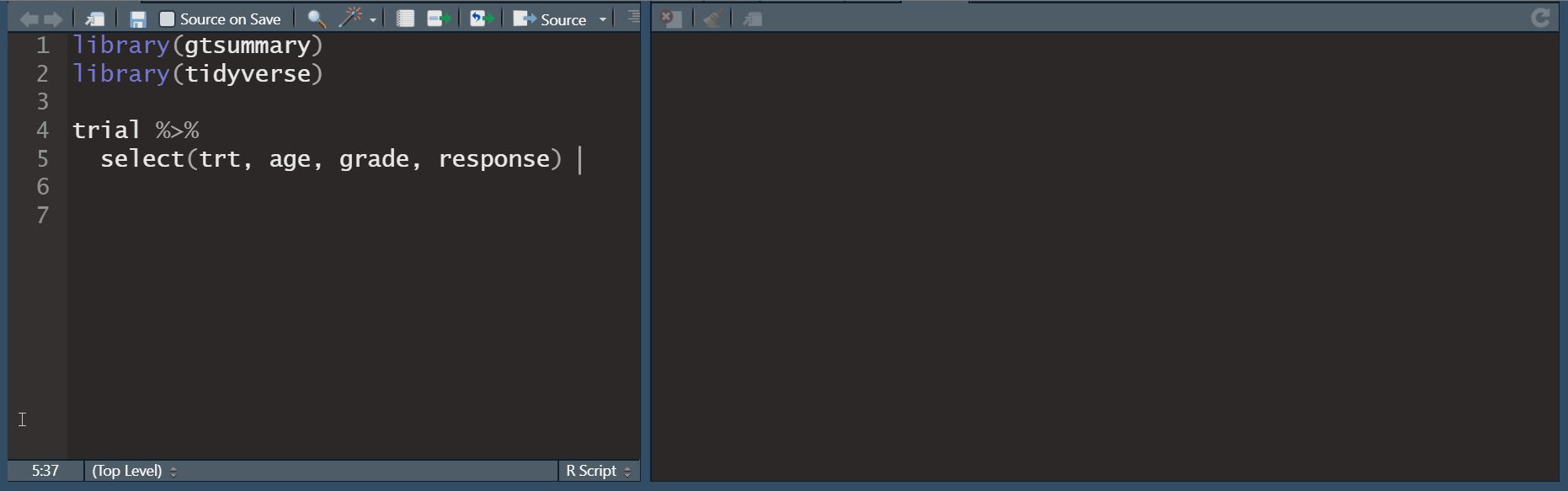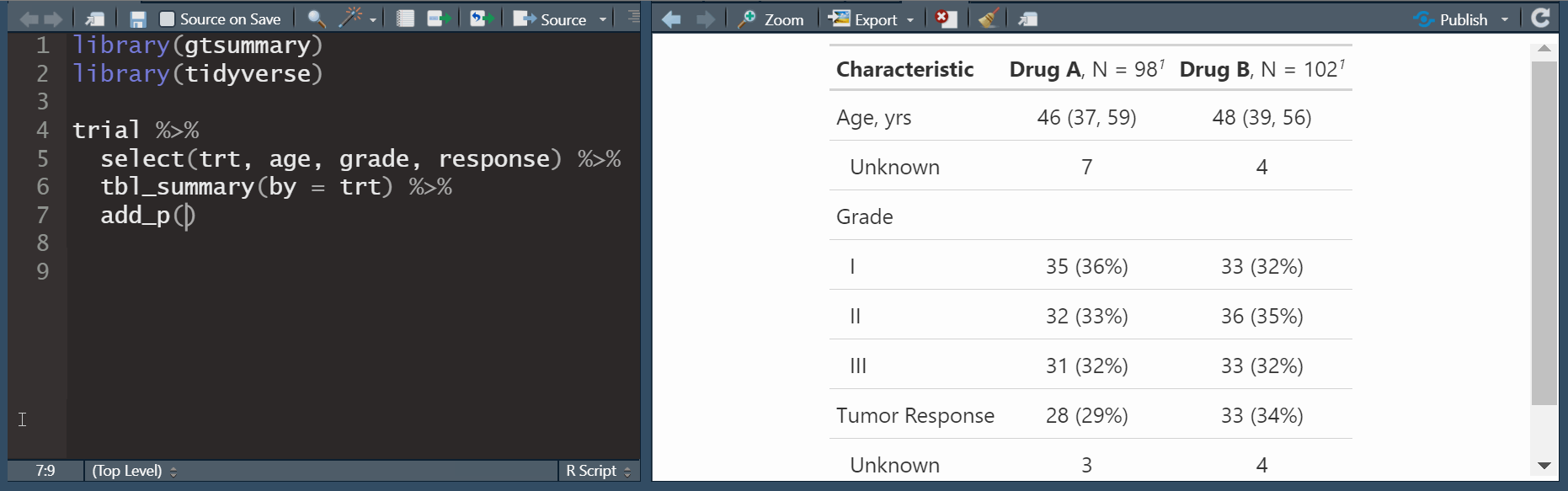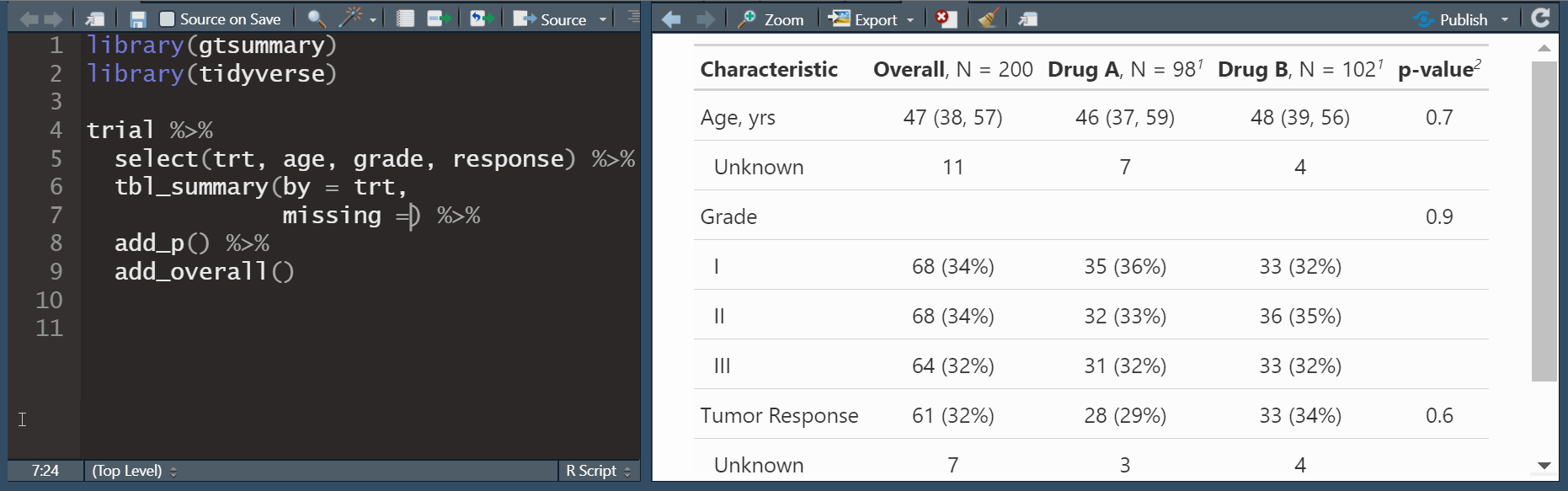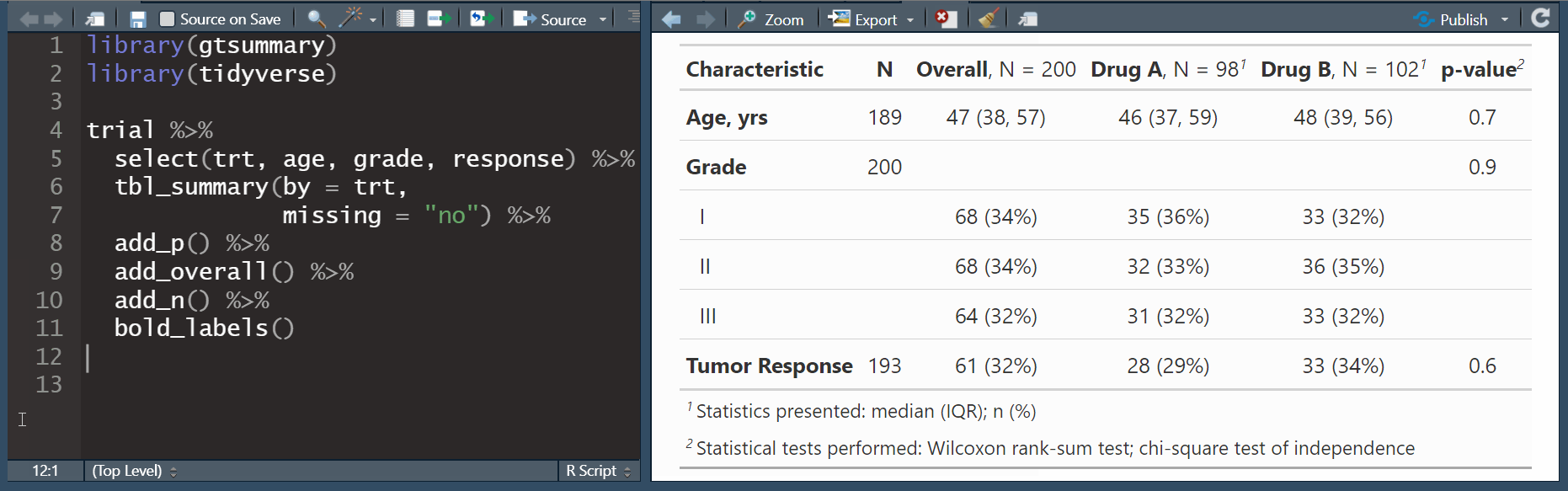
add_overall(): adds a column of overall statisticsadd_n(): adds a column with the sample size
Update tbl_summary() with add_*()
| Characteristic | N | Overall N = 200 |
Drug A N = 98 |
Drug B N = 102 |
|---|---|---|---|---|
| Age, Median (Q1, Q3) | 189 | 47 (38, 57) | 46 (37, 60) | 48 (39, 56) |
| Grade, No. (%) | 200 | |||
| I | 68 (34%) | 35 (36%) | 33 (32%) | |
| II | 68 (34%) | 32 (33%) | 36 (35%) | |
| III | 64 (32%) | 31 (32%) | 33 (32%) | |
| Tumor Response, No. (%) | 193 | 61 (32%) | 28 (29%) | 33 (34%) |
add_overall(): adds a column of overall statisticsadd_n(): adds a column with the sample sizeadd_stat_label(): adds a description of the reported statistic
Update with bold_*()/italicize_*()
| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
p-value2 |
|---|---|---|---|
| Age | 46 (37, 60) | 48 (39, 56) | 0.7 |
| Unknown | 7 | 4 | |
| Grade | 0.9 | ||
| I | 35 (36%) | 33 (32%) | |
| II | 32 (33%) | 36 (35%) | |
| III | 31 (32%) | 33 (32%) | |
| Tumor Response | 28 (29%) | 33 (34%) | 0.5 |
| Unknown | 3 | 4 | |
| 1 Median (Q1, Q3); n (%) | |||
| 2 Wilcoxon rank sum test; Pearson’s Chi-squared test | |||
bold_labels(): bold the variable labelsitalicize_levels(): italicize the variable levelsbold_p(): bold p-values according a specified threshold
Update tbl_summary() with modify_*()
| Characteristic | Drug | |
|---|---|---|
| Group A1 | Group B1 | |
| Age | 46 (37, 60) | 48 (39, 56) |
| Grade | ||
| I | 35 (36%) | 33 (32%) |
| II | 32 (33%) | 36 (35%) |
| III | 31 (32%) | 33 (32%) |
| Tumor Response | 28 (29%) | 33 (34%) |
| 1 median (IQR) for continuous; n (%) for categorical | ||
- Use
show_header_names()to see the internal header names available for use inmodify_header()
Column names
Column Name Column Header
------------ -------------------
label **Characteristic**
stat_1 **Group A**
stat_2 **Group B** all_stat_cols() selects columns "stat_1" and "stat_2"
Update tbl_summary() with add_*()
trial |>
select(trt, marker, response) |>
tbl_summary(
by = trt,
statistic = list(marker ~ "{mean} ({sd})",
response ~ "{p}%"),
missing = "no"
) |>
add_difference()| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
Difference2 | 95% CI2,3 | p-value2 |
|---|---|---|---|---|---|
| Marker Level (ng/mL) | 1.02 (0.89) | 0.82 (0.83) | 0.20 | -0.05, 0.44 | 0.12 |
| Tumor Response | 29% | 34% | -4.2% | -18%, 9.9% | 0.6 |
| 1 Mean (SD); % | |||||
| 2 Welch Two Sample t-test; 2-sample test for equality of proportions with continuity correction | |||||
| 3 CI = Confidence Interval | |||||
add_difference(): mean and rate differences between two groups. Can also be adjusted differences
Update tbl_summary() with add_*()
Where are the ARDs?
ARDs are the backbone for all calculations in gtsummary
Every gtsummary table saves the ARDs from each calculation
They can be extracted individually, or combined.
[1] "list"[1] "tbl_summary" "add_p" group1 variable context stat_name stat_label stat
1 trt age stats_wi… estimate Median o… -1
2 trt age stats_wi… statistic X-square… 4323
3 trt age stats_wi… p.value p-value 0.718
4 trt age stats_wi… conf.low CI Lower… -5
5 trt age stats_wi… conf.high CI Upper… 4
6 trt age stats_wi… method method Wilcoxon…
7 trt age stats_wi… alternative alternat… two.sided
8 trt age stats_wi… mu mu 0
9 trt age stats_wi… paired Paired t… FALSE
10 trt age stats_wi… exact exact
11 trt age stats_wi… correct correct TRUE
12 trt age stats_wi… conf.int conf.int TRUE
13 trt age stats_wi… conf.level CI Confi… 0.95
14 trt age stats_wi… tol.root tol.root 0
15 trt age stats_wi… digits.rank digits.r… InfAdd-on functions in {gtsummary}
And many more!
See the documentation at http://www.danieldsjoberg.com/gtsummary/reference/index.html
And a detailed tbl_summary() vignette at http://www.danieldsjoberg.com/gtsummary/articles/tbl_summary.html
{gtsummary} Exercise 1
Navigate to Posit Cloud script
04-gtsummary_exercise1.R.Create the table outlined in the script.
Add the “completed” sticky note to your laptop when complete.
10:00
Cross-tabulation with tbl_cross()
tbl_cross() is a wrapper for tbl_summary() for n x m tables
Continuous Summaries with tbl_continuous()
tbl_continuous() summarizes a continuous variable by 1, 2, or more categorical variables
Wide Summaries with tbl_wide_summary()
tbl_wide_summary() summarizes a continuous variable with summary statistics spread across columns
Wide Summaries with tbl_wide_summary()
| Characteristic | Median | Q1, Q3 |
|---|---|---|
| Age | 47 | 38, 57 |
| Marker Level (ng/mL) | 0.64 | 0.22, 1.41 |
Naturally, you can change the statistics, and which appear in each column.
tbl_merge()/tbl_stack()
tbl_merge() for side-by-side tables
tbl_n <-
tbl_summary(trial, include = grade, statistic = grade ~ "{n}") |>
modify_header(all_stat_cols() ~ "**N**") |> # update column header
modify_footnote(all_stat_cols() ~ NA) # remove footnote
tbl_age <-
tbl_continuous(trial, include = grade, variable = age, by = trt) |>
modify_header(all_stat_cols() ~ "**{level}**") # update header
# combine the tables side by side
list(tbl_n, tbl_age) |>
tbl_merge(tab_spanner = FALSE) # suppress default header| Characteristic | N | Drug A1 | Drug B1 |
|---|---|---|---|
| Grade | |||
| I | 68 | 46 (36, 60) | 48 (42, 55) |
| II | 68 | 45 (31, 55) | 51 (42, 58) |
| III | 64 | 52 (42, 61) | 45 (36, 52) |
| 1 Age: Median (Q1, Q3) | |||
tbl_stack() to combine vertically
tbl_drug_a <- trial |>
dplyr::filter(trt == "Drug A") |>
tbl_summary(include = c(response, death), missing = "no")
tbl_drug_b <- trial |>
dplyr::filter(trt == "Drug B") |>
tbl_summary(include = c(response, death), missing = "no")
# stack the two tables
list(tbl_drug_a, tbl_drug_b) |>
tbl_stack(group_header = c("Drug A", "Drug B")) |> # optionally include headers for each table
modify_header(all_stat_cols() ~ "**Outcome Rates**")| Characteristic | Outcome Rates1 |
|---|---|
| Drug A | |
| Tumor Response | 28 (29%) |
| Patient Died | 52 (53%) |
| Drug B | |
| Tumor Response | 33 (34%) |
| Patient Died | 60 (59%) |
| 1 n (%) | |
tbl_strata() for stratified tables
| Characteristic | Drug A | Drug B | ||
|---|---|---|---|---|
| n | % | n | % | |
| Tumor Response | 28 | 29% | 33 | 34% |
| Patient Died | 52 | 53% | 60 | 59% |
The default is to combine stratified tables with tbl_merge().
tbl_strata() for stratified tables
We can also stack the tables.
Define custom function tbl_cmh()

Define custom function tbl_cmh()

Cobbling Tables Together
Most of the tables we create in the pharma space come from a catalog of standard tables.
Custom or one-off tables are often quite difficult and time intensive to create.
The {gtsummary} package makes it simple to break complex tables into their simple parts and cobble them together in the end.
Moreover, the internal structure of a gtsummary table is super simple: a data frame and instructions on how to print that data frame to make it cute. If needed, you can directly modify the underlying data frame with
modify_table_body().
# A tibble: 6 × 7
variable var_type row_type var_label label stat_1 stat_2
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 age continuous label Age Age 46 (37, 60) 48 (39, 56)
2 age continuous missing Age Unknown 7 4
3 grade categorical label Grade Grade <NA> <NA>
4 grade categorical level Grade I 35 (36%) 33 (32%)
5 grade categorical level Grade II 32 (33%) 36 (35%)
6 grade categorical level Grade III 31 (32%) 33 (32%) ARD-first tables
ARD-first Tables
Similar to functions that accept a data frame, the package exports functions with nearly identical APIs that accept an ARD.
ARD-first Tables
We can use the skills we learned earlier today to create ARDs for gtsummary tables.
variable variable_level context stat_name stat_label stat
1 age continuo… N N 189
2 age continuo… mean Mean 47.238
3 age continuo… sd SD 14.312
4 age continuo… median Median 47
5 age continuo… p25 Q1 38
6 age continuo… p75 Q3 57
7 age continuo… min Min 6
8 age continuo… max Max 83
9 age missing N_obs Vector L… 200
10 age missing N_miss N Missing 11ARD-first Tables
We can simply use the ARD from the previous slide, and pass it to tbl_ard_summary() for a summary table.
ARD-first Tables
Now let’s try a somewhat more complicated table.
trial |>
labelled::set_variable_labels(age = "Age, years") |>
ard_stack(
.by = trt,
ard_continuous(
variables = age,
fmt_fn = age ~ list(sd = 2)
),
ard_categorical(variables = grade),
ard_dichotomous(variables = response),
# add these for best-looking tables
.attributes = TRUE,
.missing = TRUE
) |>
tbl_ard_summary(
by = trt,
type = all_continuous() ~ "continuous2",
statistic = all_continuous() ~ c("{mean} ({sd})", "{min} - {max}"),
missing = "no"
) |>
modify_caption("**Table 1. Subject Demographics**")ARD-first Tables
| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|
| Age, years | ||
| Mean (SD) | 47.0 (14.71) | 47.4 (14.01) |
| Min - Max | 6.0 - 78.0 | 9.0 - 83.0 |
| Grade | ||
| I | 35 (35.7%) | 33 (32.4%) |
| II | 32 (32.7%) | 36 (35.3%) |
| III | 31 (31.6%) | 33 (32.4%) |
| Tumor Response | 28 (29.5%) | 33 (33.7%) |
| 1 n (%) | ||
ARD-first Table Shells
trial |>
labelled::set_variable_labels(age = "Age, years") |>
ard_stack(
.by = trt,
ard_continuous(
variables = age,
fmt_fn = age ~ list(sd = 2)
),
ard_categorical(variables = grade),
ard_dichotomous(variables = response),
# add these for best-looking tables
.attributes = TRUE,
.missing = TRUE
) |>
dplyr::mutate(fmt_fn = list(\(x) "xx.x")) |>
tbl_ard_summary(
by = trt,
type = all_continuous() ~ "continuous2",
statistic = all_continuous() ~ c("{mean} ({sd})", "{min} - {max}"),
missing = "no"
) |>
modify_header(all_stat_cols() ~ "**{level}** \nN = xx")ARD-first Table Shells
| Characteristic | Drug A N = xx1 |
Drug B N = xx1 |
|---|---|---|
| Age, years | ||
| Mean (SD) | xx.x (xx.x) | xx.x (xx.x) |
| Min - Max | xx.x - xx.x | xx.x - xx.x |
| Grade | ||
| I | xx.x (xx.x%) | xx.x (xx.x%) |
| II | xx.x (xx.x%) | xx.x (xx.x%) |
| III | xx.x (xx.x%) | xx.x (xx.x%) |
| Tumor Response | xx.x (xx.x%) | xx.x (xx.x%) |
| 1 n (%) | ||
{gtsummary} Exercise 2
Navigate to Posit Cloud script
05-gtsummary_exercise2.R.Create the table outlined in the script.
Add the “completed” sticky note to your laptop when complete.
10:00
{gtsummary} themes
{gtsummary} theme basics
A theme is a set of customization preferences that can be easily set and reused.
Themes control default settings for existing functions
Themes control more fine-grained customization not available via arguments or helper functions
Easily use one of the available themes, or create your own
{gtsummary} default theme
{gtsummary} theme_gtsummary_journal()
| Characteristic | Drug A N = 98 |
Drug B N = 102 |
|---|---|---|
| Age, Median (IQR) | 46 (37 – 60) | 48 (39 – 56) |
| Unknown | 7 | 4 |
| Tumor Response, n (%) | 28 (29) | 33 (34) |
| Unknown | 3 | 4 |
{gtsummary} theme_gtsummary_language()
| 特色 | Drug A N = 981 |
Drug B N = 1021 |
P 值2 |
|---|---|---|---|
| Age | 46 (37, 60) | 48 (39, 56) | 0.7 |
| 未知 | 7 | 4 | |
| Tumor Response | 28 (29%) | 33 (34%) | 0.5 |
| 未知 | 3 | 4 | |
| 1 中位數 (Q1, Q3); n (%) | |||
| 2 Wilcoxon 排序和檢定; 卡方 獨立性檢定 | |||
Language options:
- German
- English
- Spanish
- French
- Gujarati
- Hindi
- Icelandic
- Japanese
- Korean
- Marathi
- Dutch
- Norwegian
- Portuguese
- Swedish
- Chinese Simplified
- Chinese Traditional
{gtsummary} theme_gtsummary_compact()
reset_gtsummary_theme()
theme_gtsummary_compact()
trial |>
tbl_summary(
by = trt,
include = c(age, response)
) |>
modify_caption("Compact Theme")| Characteristic | Drug A N = 981 |
Drug B N = 1021 |
|---|---|---|
| Age | 46 (37, 60) | 48 (39, 56) |
| Unknown | 7 | 4 |
| Tumor Response | 28 (29%) | 33 (34%) |
| Unknown | 3 | 4 |
| 1 Median (Q1, Q3); n (%) | ||
Reduces padding and font size
A pharma theme?
While not yet exported from gtsummary, we can create a theme for tables that look more like what we expect in pharma.
Fixed-width font
Continuous variable summaries default to multi-line
Function for rounding percentages includes leading white space
Default right alignment on summary statistics
| Characteristic | Placebo N = 86 |
Xanomeline Low Dose N = 84 |
Xanomeline High Dose N = 84 |
|---|---|---|---|
| Age | |||
| Median (Q1, Q3) | 76.0 (69.0, 82.0) | 77.5 (71.0, 82.0) | 76.0 (70.5, 80.0) |
| Mean (SD) | 75.2 (8.6) | 75.7 (8.3) | 74.4 (7.9) |
| Min - Max | 52.0 - 89.0 | 51.0 - 88.0 | 56.0 - 88.0 |
| Age Group, n (%) | |||
| <65 | 14 (16.3%) | 8 ( 9.5%) | 11 (13.1%) |
| 65-80 | 42 (48.8%) | 47 (56.0%) | 55 (65.5%) |
| >80 | 30 (34.9%) | 29 (34.5%) | 18 (21.4%) |
| Female, n (%) | 53 (61.6%) | 50 (59.5%) | 40 (47.6%) |
{gtsummary} set_gtsummary_theme()
set_gtsummary_theme()to use a custom theme.See the {gtsummary} + themes vignette for examples
http://www.danieldsjoberg.com/gtsummary/articles/themes.html
{gtsummary} print engines
{gtsummary} print engines

{gtsummary} print engines
Use any print engine to customize table
In Closing
{gtsummary} website
Package Authors/Contributors
Daniel D. Sjoberg
Joseph Larmarange
Michael Curry
Jessica Lavery
Karissa Whiting
Emily C. Zabor
Xing Bai
Esther Drill
Jessica Flynn
Margie Hannum
Stephanie Lobaugh
Shannon Pileggi
Amy Tin
Gustavo Zapata Wainberg
Other Contributors
@abduazizR, @ablack3, @ABohynDOE, @ABorakati, @adilsonbauhofer, @aghaynes, @ahinton-mmc, @aito123, @akarsteve, @akefley, @albamrt, @albertostefanelli, @alecbiom, @alexandrayas, @alexis-catherine, @AlexZHENGH, @alnajar, @amygimma, @anaavu, @anddis, @andrader, @Andrzej-Andrzej, @angelgar, @arbet003, @arnmayer, @aspina7, @AurelienDasre, @awcm0n, @ayogasekaram, @barretmonchka, @barthelmes, @bc-teixeira, @bcjaeger, @BeauMeche, @benediktclaus, @benwhalley, @berg-michael, @bhattmaulik, @BioYork, @blue-abdur, @brachem-christian, @brianmsm, @browne123, @bwiernik, @bx259, @calebasaraba, @CarolineXGao, @CharlyMarie, @ChongTienGoh, @Chris-M-P, @chrisleitzinger, @cjprobst, @ClaudiaCampani, @clmawhorter, @CodieMonster, @coeusanalytics, @coreysparks, @CorradoLanera, @crystalluckett-sanofi, @ctlamb, @dafxy, @DanChaltiel, @DanielPark-MGH, @davideyre, @davidgohel, @davidkane9, @DavisVaughan, @dax44, @dchiu911, @ddsjoberg, @DeFilippis, @denis-or, @dereksonderegger, @derekstein, @DesiQuintans, @dieuv0, @dimbage, @discoleo, @djbirke, @dmenne, @DrDinhLuong, @edelarua, @edrill, @Eduardo-Auer, @ElfatihHasabo, @emilyvertosick, @eokoshi, @ercbk, @eremingt, @erikvona, @eugenividal, @eweisbrod, @fdehrich, @feizhadj, @fh-jsnider, @fh-mthomson, @FrancoisGhesquiere, @ge-generation, @Generalized, @ghost, @giorgioluciano, @giovannitinervia9, @gjones1219, @gorkang, @GuiMarthe, @gungorMetehan, @hass91, @hescalar, @HichemLa, @hichew22, @hr70, @huftis, @hughjonesd, @iaingallagher, @ilyamusabirov, @IndrajeetPatil, @irene9116, @IsadoraBM, @j-tamad, @jalavery, @jaromilfrossard, @JBarsotti, @jbtov, @jeanmanguy, @jemus42, @jenifav, @jennybc, @JeremyPasco, @jerrodanzalone, @JesseRop, @jflynn264, @jhchou, @jhelvy, @jhk0530, @jjallaire, @jkylearmstrong, @jmbarajas, @jmbarbone, @JoanneF1229, @joelgautschi, @johnryan412, @JohnSodling, @jonasrekdalmathisen, @JonGretar, @jordan49er, @jsavinc, @jthomasmock, @juseer, @jwilliman, @karissawhiting, @karl-an, @kendonB, @kentm4, @klh281, @kmdono02, @kristyrobledo, @kwakuduahc1, @lamberp6, @lamhine, @larmarange, @ledermanr, @leejasme, @leslem, @levossen, @lngdet, @longjp, @lorenzoFabbri, @loukesio, @love520lfh, @lspeetluk, @ltin1214, @ltj-github, @lucavd, @LucyMcGowan, @LuiNov, @lukejenner6, @maciekbanas, @maia-sh, @malcolmbarrett, @mariamaseng, @Marsus1972, @martsobm, @Mathicaa, @matthieu-faron, @maxanes, @mayazadok2, @mbac, @mdidish, @medewitt, @meenakshi-kushwaha, @melindahiggins2000, @MelissaAssel, @Melkiades, @mfansler, @michaelcurry1123, @mikemazzucco, @mlamias, @mljaniczek, @moleps, @monitoringhsd, @motocci, @mrmvergeer, @msberends, @mvuorre, @myamortor, @myensr, @MyKo101, @nalimilan, @ndunnewind, @nikostr, @ningyile, @O16789, @oliviercailloux, @oranwutang, @palantre, @parmsam, @Pascal-Schmidt, @PaulC91, @paulduf, @pedersebastian, @perlatex, @pgseye, @philippemichel, @philsf, @polc1410, @Polperobis, @postgres-newbie, @proshano, @raphidoc, @RaviBot, @rawand-hanna, @rbcavanaugh, @remlapmot, @rich-iannone, @RiversPharmD, @rmgpanw, @roaldarbol, @roman2023, @ryzhu75, @s-j-choi, @sachijay, @saifelayan, @sammo3182, @samrodgersmelnick, @samuele-mercan, @sandhyapc, @sbalci, @sda030, @shah-in-boots, @shannonpileggi, @shaunporwal, @shengchaohou, @ShixiangWang, @simonpcouch, @slb2240, @slobaugh, @spiralparagon, @Spring75xx, @StaffanBetner, @steenharsted, @stenw, @Stephonomon, @storopoli, @stratopopolis, @strengejacke, @szimmer, @tamytsujimoto, @TAOS25, @TarJae, @themichjam, @THIB20, @tibirkrajc, @tjmeyers, @tldrcharlene, @tormodb, @toshifumikuroda, @TPDeramus, @UAB-BST-680, @uakimix, @uriahf, @Valja64, @viola-hilbert, @violet-nova, @vvm02, @will-gt, @xkcococo, @xtimbeau, @yatirbe, @yihunzeleke, @yonicd, @yoursdearboy, @YousufMohammed2002, @yuryzablotski, @zabore, @zachariae, @zaddyzad, @zawkzaw, @zdz2101, @zeyunlu, @zhangkaicr, @zhaohongxin0, @zheer-kejlberg, @zhengnow, @zhonghua723, @zlkrvsm, @zongell-star, and @Zoulf001.
Thank you
Ask on stackoverflow.com
Use the gtsummary tag
Thousands of posts!
More on ARDs at the 2024 R/Pharma Workshop
